1PSR
 
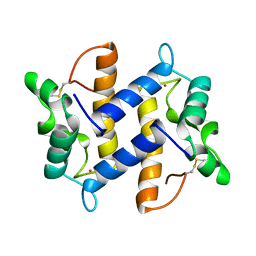 | | HUMAN PSORIASIN (S100A7) | | Descriptor: | HOLMIUM ATOM, PSORIASIN | | Authors: | Brodersen, D.E, Etzerodt, M, Madsen, P, Celis, J, Thoegersen, H.C, Nyborg, J, Kjeldgaard, M. | | Deposit date: | 1997-11-27 | | Release date: | 1999-01-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | EF-hands at atomic resolution: the structure of human psoriasin (S100A7) solved by MAD phasing.
Structure, 6, 1998
|
|
1PO5
 
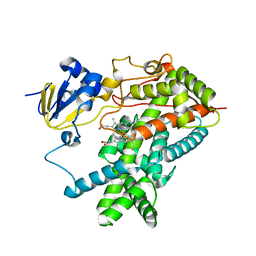 | | Structure of mammalian cytochrome P450 2B4 | | Descriptor: | Cytochrome P450 2B4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Scott, E.E, He, Y.A, Wester, M.R, White, M.A, Chin, C.C, Halpert, J.R, Johnson, E.F, Stout, C.D. | | Deposit date: | 2003-06-13 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An open conformation of mammalian cytochrome P450 2B4 at 1.6 A resolution
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1DUQ
 
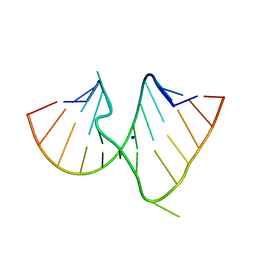 | |
2JDN
 
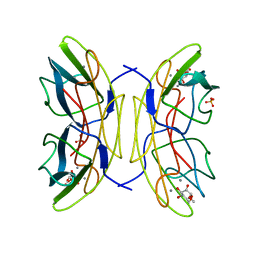 | | Mutant (S22A) of Pseudomonas aeruginosa lectin II (PA-IIL) complexed with methyl-a-L-mannopyranoside | | Descriptor: | CALCIUM ION, FUCOSE-BINDING LECTIN PA-IIL, SULFATE ION, ... | | Authors: | Adam, J, Pokorna, M, Sabin, C, Mitchell, E.P, Imberty, A, Wimmerova, M. | | Deposit date: | 2007-01-11 | | Release date: | 2007-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Engineering of Pa-Iil Lectin from Pseudomonas Aeruginosa - Unravelling the Role of the Specificity Loop for Sugar Preference.
Bmc Struct.Biol., 7, 2007
|
|
2JDU
 
 | | Mutant (G24N) of Pseudomonas aeruginosa lectin II (PA-IIL) complexed with methyl-a-L-fucopyranoside | | Descriptor: | CALCIUM ION, FUCOSE-BINDING LECTIN PA-IIL, GLYCEROL, ... | | Authors: | Adam, J, Pokorna, M, Sabin, C, Mitchell, E.P, Imberty, A, Wimmerova, M. | | Deposit date: | 2007-01-12 | | Release date: | 2007-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Engineering of Pa-Iil Lectin from Pseudomonas Aeruginosa - Unravelling the Role of the Specificity Loop for Sugar Preference.
Bmc Struct.Biol., 7, 2007
|
|
2JDY
 
 | | Mutant (G24N) of Pseudomonas aeruginosa lectin II (PA-IIL) complexed with methyl-b-D-mannoyranoside | | Descriptor: | CALCIUM ION, FUCOSE-BINDING LECTIN PA-IIL, methyl alpha-D-mannopyranoside | | Authors: | Adam, J, Pokorna, M, Sabin, C, Mitchell, E.P, Imberty, A, Wimmerova, M. | | Deposit date: | 2007-01-12 | | Release date: | 2007-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Engineering of Pa-Iil Lectin from Pseudomonas Aeruginosa - Unravelling the Role of the Specificity Loop for Sugar Preference.
Bmc Struct.Biol., 7, 2007
|
|
2JDP
 
 | | Mutant (S23A) of Pseudomonas aeruginosa lectin II (PA-IIL) complexed with methyl-a-L-fucopyranoside | | Descriptor: | CALCIUM ION, FUCOSE-BINDING LECTIN PA-IIL, methyl alpha-L-fucopyranoside | | Authors: | Adam, J, Pokorna, M, Sabin, C, Mitchell, E.P, Imberty, A, Wimmerova, M. | | Deposit date: | 2007-01-11 | | Release date: | 2007-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Engineering of Pa-Iil Lectin from Pseudomonas Aeruginosa - Unravelling the Role of the Specificity Loop for Sugar Preference.
Bmc Struct.Biol., 7, 2007
|
|
7ZAV
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3 | | Authors: | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | Deposit date: | 2022-03-22 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
1BEN
 
 | | INSULIN COMPLEXED WITH 4-HYDROXYBENZAMIDE | | Descriptor: | 4-HYDROXYBENZAMIDE, CHLORIDE ION, HUMAN INSULIN, ... | | Authors: | Smith, G.D, Ciszak, E, Pangborn, W. | | Deposit date: | 1996-02-15 | | Release date: | 1996-07-11 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A novel complex of a phenolic derivative with insulin: structural features related to the T-->R transition.
Protein Sci., 5, 1996
|
|
2AAI
 
 | | Crystallographic refinement of ricin to 2.5 Angstroms | | Descriptor: | RICIN (A CHAIN), RICIN (B CHAIN), alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rutenber, E, Katzin, B.J, Montfort, W, Villafranca, J.E, Ernst, S.R, Collins, E.J, Mlsna, D, Monzingo, A.F, Ready, M.P, Robertus, J.D. | | Deposit date: | 1993-09-07 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic refinement of ricin to 2.5 A.
Proteins, 10, 1991
|
|
2A0Q
 
 | | Structure of thrombin in 400 mM potassium chloride | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Papaconstantinou, M, Carrell, C.J, Pineda, A.O, Bobofchak, K.M, Mathews, F.S, Flordellis, C.S, Maragoudakis, M.E, Tsopanoglou, N.E, di Cera, E. | | Deposit date: | 2005-06-16 | | Release date: | 2005-07-12 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Thrombin Functions through Its RGD Sequence in a Non-canonical Conformation.
J.Biol.Chem., 280, 2005
|
|
1E1Q
 
 | | BOVINE MITOCHONDRIAL F1-ATPASE AT 100K | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BOVINE MITOCHONDRIAL F1-ATPASE, MAGNESIUM ION, ... | | Authors: | Braig, K, Menz, R.I, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2000-05-10 | | Release date: | 2000-06-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structure of Bovine Mitochondrial F1-ATPase Inhibited by Mg2+Adp and Aluminium Fluoride
Structure, 8, 2000
|
|
2HEO
 
 | | General Structure-Based Approach to the Design of Protein Ligands: Application to the Design of Kv1.2 Potassium Channel Blockers. | | Descriptor: | 5'-D(*TP*CP*GP*CP*GP*CP*G)-3', Z-DNA binding protein 1 | | Authors: | Magis, C, Gasparini, S, Charbonnier, J.B, Stura, E, Le Du, M.H, Menez, A, Cuniasse, P. | | Deposit date: | 2006-06-21 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-based secondary structure-independent approach to design protein ligands: Application to the design of Kv1.2 potassium channel blockers.
J.Am.Chem.Soc., 128, 2006
|
|
1Y33
 
 | | Crystal structure of the complex of subtilisin BPN' with chymotrypsin inhibitor 2 T58P mutant | | Descriptor: | CALCIUM ION, CITRIC ACID, POLYETHYLENE GLYCOL (N=34), ... | | Authors: | Radisky, E.S, Lu, C.J, Kwan, G, Koshland Jr, D.E. | | Deposit date: | 2004-11-23 | | Release date: | 2005-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of the intramolecular hydrogen bond network in the inhibitory power of chymotrypsin inhibitor 2
Biochemistry, 44, 2005
|
|
2HG4
 
 | | Structure of the ketosynthase-acyltransferase didomain of module 5 from DEBS. | | Descriptor: | 6-Deoxyerythronolide B Synthase, ACETATE ION, CHLORIDE ION, ... | | Authors: | Tang, Y, Kim, C.Y, Mathews, I.I, Cane, D.E, Khosla, C. | | Deposit date: | 2006-06-26 | | Release date: | 2006-07-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | The 2.7-A crystal structure of a 194-kDa homodimeric fragment of the 6-deoxyerythronolide B synthase.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2OYQ
 
 | | Crystal structure of RB69 gp43 in complex with DNA with 5-NIMP opposite an abasic site analog | | Descriptor: | 1-{2-DEOXY-5-O-[(R)-HYDROXY{[(R)-HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}PHOSPHORYL]-BETA-D-ERYTHRO-PENTOFURANOSYL}-5-NITRO -1H-INDOLE, DNA polymerase, MAGNESIUM ION, ... | | Authors: | Zahn, K.E, Belrhali, H, Wallace, S.S, Doublie, S. | | Deposit date: | 2007-02-22 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Caught bending the a-rule: crystal structures of translesion DNA synthesis with a non-natural nucleotide.
Biochemistry, 46, 2007
|
|
1JWL
 
 | | Structure of the Dimeric lac Repressor/Operator O1/ONPF Complex | | Descriptor: | 2-nitrophenyl beta-D-fucopyranoside, 5'-D(*AP*GP*AP*AP*T*TP*GP*TP*GP*AP*GP*CP*GP*GP*AP*TP*AP*AP*CP*AP*AP*TP*T)-3', 5'-D(*TP*AP*AP*TP*TP*GP*TP*TP*AP*TP*CP*CP*GP*CP*TP*CP*AP*CP*AP*AP*TP*TP*C)-3', ... | | Authors: | Bell, C.E, Lewis, M. | | Deposit date: | 2001-09-04 | | Release date: | 2001-10-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Crystallographic analysis of Lac repressor bound to natural operator O1.
J.Mol.Biol., 312, 2001
|
|
3QOQ
 
 | |
4PQU
 
 | | Crystal structure of HIV-1 Reverse Transcriptase in complex with RNA/DNA and dATP | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 5'-D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*G)-3', ... | | Authors: | Das, K, Bandwar, R.P, Arnold, E. | | Deposit date: | 2014-03-04 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.508 Å) | | Cite: | Structures of HIV-1 RT-RNA/DNA ternary complexes with dATP and nevirapine reveal conformational flexibility of RNA/DNA: insights into requirements for RNase H cleavage.
Nucleic Acids Res., 42, 2014
|
|
1TF3
 
 | | TFIIIA FINGER 1-3 BOUND TO DNA, NMR, 22 STRUCTURES | | Descriptor: | 5S RNA GENE, TRANSCRIPTION FACTOR IIIA, ZINC ION | | Authors: | Foster, M.P, Wuttke, D.S, Radhakrishnan, I, Case, D.A, Gottesfeld, J.M, Wright, P.E. | | Deposit date: | 1997-07-01 | | Release date: | 1997-09-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Domain packing and dynamics in the DNA complex of the N-terminal zinc fingers of TFIIIA.
Nat.Struct.Biol., 4, 1997
|
|
1E1R
 
 | | BOVINE MITOCHONDRIAL F1-ATPASE INHIBITED BY MG2+ADP AND ALUMINIUM FLUORIDE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, BOVINE MITOCHONDRIAL F1-ATPASE, ... | | Authors: | Braig, K, Menz, R.I, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2000-05-10 | | Release date: | 2000-06-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Bovine Mitochondrial F1-ATPase Inhibited by Mg2+Adp and Aluminium Fluoride
Structure, 8, 2000
|
|
1EGW
 
 | | CRYSTAL STRUCTURE OF MEF2A CORE BOUND TO DNA | | Descriptor: | DNA (5'-D(*AP*AP*AP*GP*CP*TP*AP*TP*TP*AP*TP*TP*AP*GP*CP*TP*T)-3'), DNA (5'-D(*TP*AP*AP*GP*CP*TP*AP*AP*TP*AP*AP*TP*AP*GP*CP*TP*T)-3'), MADS BOX TRANSCRIPTION ENHANCER FACTOR 2, ... | | Authors: | Santelli, E, Richmond, T.J. | | Deposit date: | 2000-02-17 | | Release date: | 2000-03-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of MEF2A core bound to DNA at 1.5 A resolution.
J.Mol.Biol., 297, 2000
|
|
2LTQ
 
 | | High resolution structure of DsbB C41S by joint calculation with solid-state NMR and X-ray data | | Descriptor: | Disulfide bond formation protein B, Fab fragment heavy chain, Fab fragment light chain, ... | | Authors: | Tang, M, Sperling, L.J, Schwieters, C.D, Nesbitt, A.E, Gennis, R.B, Rienstra, C.M. | | Deposit date: | 2012-05-30 | | Release date: | 2013-02-27 | | Last modified: | 2023-06-14 | | Method: | SOLID-STATE NMR | | Cite: | Structure of the Disulfide Bond Generating Membrane Protein DsbB in the Lipid Bilayer.
J.Mol.Biol., 425, 2013
|
|
8U53
 
 | | Mechanically activated ion channel OSCA3.1 in nanodiscs | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CSC1-like protein ERD4, PALMITIC ACID, ... | | Authors: | Jojoa-Cruz, S, Lee, W.H, Ward, A.B. | | Deposit date: | 2023-09-12 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure-guided mutagenesis of OSCAs reveals differential activation to mechanical stimuli.
Elife, 12, 2024
|
|
8VYT
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (L372S, L536S) in complex with k-411 | | Descriptor: | 4,4'-[(1R,4R,5S)-5-(2,3-dihydro-1H-indole-1-sulfonyl)-7-oxabicyclo[2.2.1]hept-2-ene-2,3-diyl]diphenol, Estrogen receptor | | Authors: | Min, C.K, Nwachukwu, J.C, Hou, Y, Russo, R.J, Papa, A, Min, J, Peng, R, Kim, S.H, Ziegler, Y, Rangarajan, E.S, Izard, T, Katzenellenbogen, B.S, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2024-02-09 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Asymmetric allostery in estrogen receptor-alpha homodimers drives responses to the ensemble of estrogens in the hormonal milieu.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
