6TF7
 
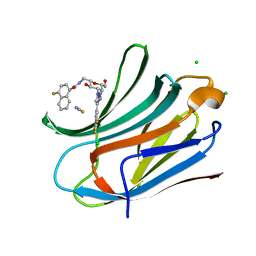 | | Human galectin-3c in complex with a galactose derivative | | Descriptor: | 4-fluoranyl-~{N}-[[(2~{S},3~{R},4~{R},5~{R},6~{R})-6-(hydroxymethyl)-3,5-bis(oxidanyl)-4-[4-[3,4,5-tris(fluoranyl)phenyl]-1,2,3-triazol-1-yl]oxan-2-yl]methyl]naphthalene-1-carboxamide, CHLORIDE ION, Galectin-3, ... | | Authors: | Nilsson, U.J, Zetterberg, F, Hakansson, M, Logan, D.T. | | Deposit date: | 2019-11-13 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | 3-Substituted 1-Naphthamidomethyl-C-galactosyls Interact with Two Unique Sub-sites for High-Affinity and High-Selectivity Inhibition of Galectin-3.
Molecules, 24, 2019
|
|
6TOM
 
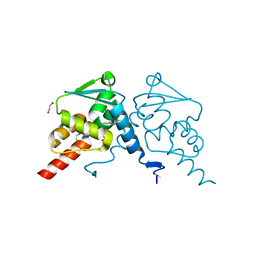 | | Crystal structure of human BCL6 BTB domain in complex with compound 1 | | Descriptor: | 1,2-ETHANEDIOL, 5-[[2-[(3~{S},5~{R})-4,4-bis(fluoranyl)-3,5-dimethyl-piperidin-1-yl]-5-chloranyl-pyrimidin-4-yl]amino]-1-methyl-3-(3-methyl-3-oxidanyl-butyl)benzimidazol-2-one, B-cell lymphoma 6 protein | | Authors: | Rodrigues, M.J, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2019-12-11 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | AchievingIn VivoTarget Depletion through the Discovery and Optimization of Benzimidazolone BCL6 Degraders.
J.Med.Chem., 63, 2020
|
|
6FZW
 
 | |
5B15
 
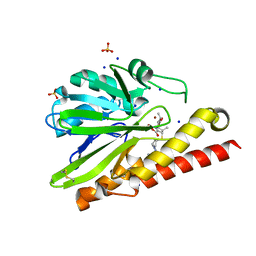 | | Crystal Structure of Metallo-beta-Lactamase SMB-1 Bound to Hydrolyzed Doripenem | | Descriptor: | (2~{S},3~{R},4~{S})-2-[(2~{S},3~{R})-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-3-methyl-4-[(3~{S},5~{S})-5-[(sulfamoylamino)methyl]pyrrolidin-3-yl]sulfanyl-3,4-dihydro-2~{H}-pyrrole-5-carboxylic acid, Metallo-beta-lactamase, SODIUM ION, ... | | Authors: | Wachino, J, Arakawa, Y. | | Deposit date: | 2015-11-23 | | Release date: | 2016-05-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Crystal Structure of Metallo-beta-Lactamase SMB-1 Bound to Hydrolyzed Doripenem
To Be Published
|
|
9Q87
 
 | | Principles of ion binding to RNA inferred from the analysis of a 1.55 Angstrom resolution bacterial ribosome structure - Part I: Mg2+ | | Descriptor: | 16S rRNA, 23S rRNA, 5S rRNA, ... | | Authors: | Leonarski, f, Henning-Knechtel, A, Kirmizialtin, S, Ennifar, E, Auffinger, P. | | Deposit date: | 2025-02-23 | | Release date: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (1.55 Å) | | Cite: | Principles of ion binding to RNA inferred from the analysis of a 1.55 Angs. resolution bacterial ribosome structure - Part I: Mg2+.
Nucleic Acids Res, 2024
|
|
5KM8
 
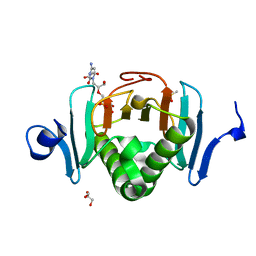 | | Human Histidine Triad Nucleotide Binding Protein 2 (hHint2) Cidofovir complex | | Descriptor: | ({[(2S)-1-(4-amino-2-oxopyrimidin-1(2H)-yl)-3-hydroxypropan-2-yl]oxy}methyl)phosphonic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Maize, K.M, Finzel, B.C. | | Deposit date: | 2016-06-26 | | Release date: | 2017-06-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Crystal Structure Based Guide to the Design of Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) Activated ProTides.
Mol. Pharm., 14, 2017
|
|
6BSC
 
 | |
6IC6
 
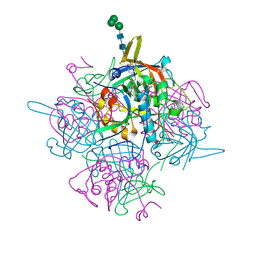 | | Human cathepsin-C in complex with cyclopropyl peptidyl nitrile inhibitor 1 | | Descriptor: | (2~{S})-~{N}-[(1~{R},2~{R})-1-(aminomethyl)-2-[4-[4-(trifluoromethyl)phenyl]phenyl]cyclopropyl]-2-azanyl-butanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Hakansson, M, Logan, D.T, Korkmaz, B, Lesner, A, Wysocka, M, Gieldon, A, Gauthier, F, Jenne, D, Lauritzen, C, Pedersen, J. | | Deposit date: | 2018-12-02 | | Release date: | 2019-04-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Structure-based design and in vivo anti-arthritic activity evaluation of a potent dipeptidyl cyclopropyl nitrile inhibitor of cathepsin C.
Biochem. Pharmacol., 164, 2019
|
|
7Y52
 
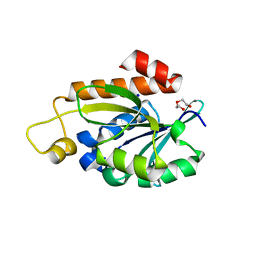 | | Crystal structure of peptidyl-tRNA hydrolase from Enterococcus faecium | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Peptidyl-tRNA hydrolase, ... | | Authors: | Pandey, R, Zohib, M, Mundra, S, Pal, R.K, Arora, A. | | Deposit date: | 2022-06-16 | | Release date: | 2024-01-17 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Characterization of structure of peptidyl-tRNA hydrolase from Enterococcus faecium and its inhibition by a pyrrolinone compound.
Int.J.Biol.Macromol., 275, 2024
|
|
5CKQ
 
 | | CUB1-EGF-CUB2 domains of rat MASP-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Mannan-binding lectin serine protease 1, ... | | Authors: | Nan, R, Furze, C.M, Wright, D.W, Gor, J, Wallis, R, Perkins, S.J. | | Deposit date: | 2015-07-15 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.704 Å) | | Cite: | Flexibility in Mannan-Binding Lectin-Associated Serine Proteases-1 and -2 Provides Insight on Lectin Pathway Activation.
Structure, 25, 2017
|
|
5BMP
 
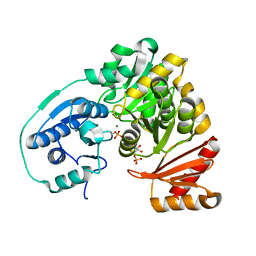 | |
9HSR
 
 | |
5BWT
 
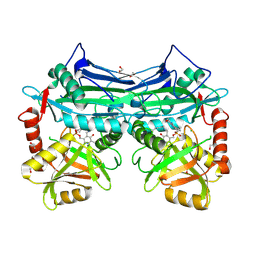 | |
1Y9W
 
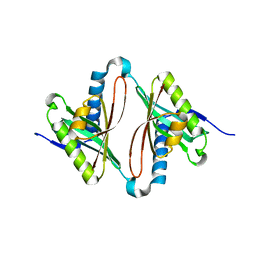 | | Structural Genomics, 1.9A crystal structure of an acetyltransferase from Bacillus cereus ATCC 14579 | | Descriptor: | Acetyltransferase | | Authors: | Zhang, R, Li, H, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-12-16 | | Release date: | 2005-02-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9A crystal structure of an acetyltransferase from Bacillus cereus ATCC 14579
To be Published, 2005
|
|
2W41
 
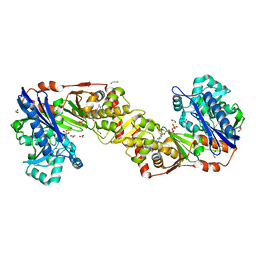 | | Crystal structure of Plasmodium falciparum glycerol kinase with ADP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL KINASE, ... | | Authors: | Schnick, C, Polley, S.D, Fivelman, Q.L, Ranford-Cartwright, L, Wilkinson, S.R, Brannigan, J.A, Wilkinson, A.J, Baker, D.A. | | Deposit date: | 2008-11-18 | | Release date: | 2008-12-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structure and Non-Essential Function of Glycerol Kinase in Plasmodium Falciparum Blood Stages.
Mol.Microbiol., 71, 2009
|
|
5GK3
 
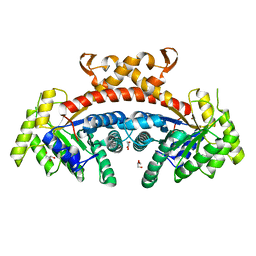 | | Native structure of fructose 1,6-bisphosphate aldolase from Escherichia coli at 1.8 Angstrom resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, Fructose-bisphosphate aldolase class 2, GLYCEROL, ... | | Authors: | Tran, T.H, Huynh, K.H, Ho, T.H, Kang, L.W. | | Deposit date: | 2016-07-03 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Apo structure of fructose 1,6-bisphosphate aldolase from Escherichia coli at 1.8 Angstrom resolution
To Be Published
|
|
5AYT
 
 | | Crystal structure of transthyretin in complex with L6 | | Descriptor: | 2-oxidanyl-6-(phenylcarbonyl)benzo[de]isoquinoline-1,3-dione, Transthyretin | | Authors: | Yokoyama, T, Mizuguchi, M, Kai, H. | | Deposit date: | 2015-09-03 | | Release date: | 2016-01-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural stabilization of transthyretin by a new compound, 6-benzoyl-2-hydroxy-1H-benzo[de]isoquinoline-1,3(2H)-dione
J. Pharmacol. Sci., 129, 2015
|
|
5B18
 
 | | Crystal Structure of a Darunavir Resistant HIV-1 Protease | | Descriptor: | ACETATE ION, CHLORIDE ION, Protease | | Authors: | Suzuki, K, Ode, H, Nakashima, M, Sugiura, W, Watanabe, N, Suzuki, A, Iwatani, Y. | | Deposit date: | 2015-11-30 | | Release date: | 2016-04-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Unique Flap Conformation in an HIV-1 Protease with High-Level Darunavir Resistance
Front Microbiol, 7, 2016
|
|
6BSV
 
 | | Crystal structure of Xyloglucan Xylosyltransferase binary form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MANGANESE (II) ION, NITRATE ION, ... | | Authors: | Zabotina, O.A, Culbertson, A.T, Ehrlich, J.J, Choe, J, Honzatko, R.B. | | Deposit date: | 2017-12-04 | | Release date: | 2018-05-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.433 Å) | | Cite: | Structure of xyloglucan xylosyltransferase 1 reveals simple steric rules that define biological patterns of xyloglucan polymers.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5C8K
 
 | | EGFR kinase domain mutant "TMLR" with compound 1 | | Descriptor: | 1-cyclopentyl-N-[2-(4-methoxypiperidin-1-yl)pyrimidin-4-yl]-1H-imidazo[4,5-c]pyridin-6-amine, Epidermal growth factor receptor | | Authors: | Eigenbrot, C, Yu, C. | | Deposit date: | 2015-06-25 | | Release date: | 2015-10-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Noncovalent Mutant Selective Epidermal Growth Factor Receptor Inhibitors: A Lead Optimization Case Study.
J.Med.Chem., 58, 2015
|
|
7FWG
 
 | | Crystal Structure of human FABP4 in complex with 1-[(4-chloro-3-phenoxyphenyl)methyl]-4-hydroxypyridin-2-one, i.e. SMILES c1(cc(c(cc1)Cl)Oc1ccccc1)CN1C(=O)C=C(C=C1)O with IC50=0.319 microM | | Descriptor: | 1-[(4-chloro-3-phenoxyphenyl)methyl]-4-hydroxypyridin-2(1H)-one, Fatty acid-binding protein, adipocyte, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Obst-Sander, U, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
5VGC
 
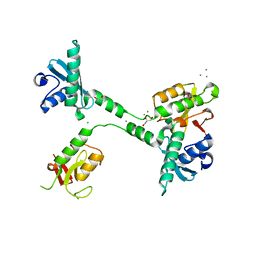 | | Crystal structure of the NleG5-1 effector (C200A) from Escherichia coli O157:H7 str. Sakai | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Borek, D, Valleau, D, Skarina, T, Jobin, M.C, Wawrzak, Z, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-04-10 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the NleG5-1 effector (C200A) from Escherichia coli O157:H7 str. Sakai
To Be Published
|
|
6BSB
 
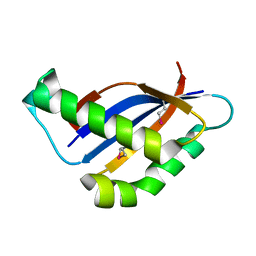 | |
4TTR
 
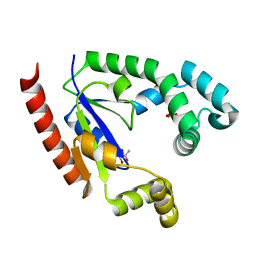 | |
7FYE
 
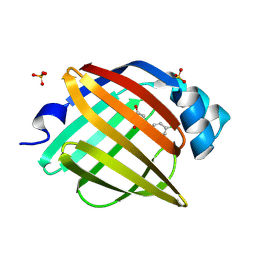 | | Crystal Structure of human FABP4 in complex with 1-[(4-chlorophenyl)methyl]-4-hydroxypyridin-2-one, i.e. SMILES C1(=CC(=O)N(C=C1)Cc1ccc(cc1)Cl)O with IC50=7.8 microM | | Descriptor: | 1-[(4-chlorophenyl)methyl]-4-hydroxypyridin-2(1H)-one, Fatty acid-binding protein, adipocyte, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Obst-Sander, U, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
