8VD2
 
 | | Human TCR ET650-4 in complex with DQ8-InsC8-15-IAPP1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hybrid insulin peptide (HIP; InsC8-15-IAPP23-29 ), MHC class II HLA-DQ-alpha chain, ... | | Authors: | Tran, T.M, Lim, J.J, Loh, T.Y, Mannering, I.S, Rossjohn, J, Reid, H.H. | | Deposit date: | 2023-12-14 | | Release date: | 2024-08-07 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A structural basis of T cell cross-reactivity to native and spliced self-antigens presented by HLA-DQ8.
J.Biol.Chem., 300, 2024
|
|
8VD0
 
 | | Human TCR ET650-4 in complex with DQ8-InsC8-15-IAPP2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Hybrid insulin peptide (HIP; InsC8-15-IAPP74-80),MHC class II HLA-DQ-beta-1 chimera, ... | | Authors: | Tran, T.M, Lim, J.J, Loh, T.Y, Mannering, I.S, Rossjohn, J, Reid, H.H. | | Deposit date: | 2023-12-14 | | Release date: | 2024-08-07 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A structural basis of T cell cross-reactivity to native and spliced self-antigens presented by HLA-DQ8.
J.Biol.Chem., 300, 2024
|
|
8VCY
 
 | | Human TCR A2.13 in complex with DQ8-InsC8-15NPY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Hybrid insulin peptide (HIP; InsC8-15-NPY68-74), ... | | Authors: | Tran, T.M, Lim, J.J, Loh, T.Y, Mannering, I.S, Rossjohn, J, Reid, H.H. | | Deposit date: | 2023-12-14 | | Release date: | 2024-08-07 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A structural basis of T cell cross-reactivity to native and spliced self-antigens presented by HLA-DQ8.
J.Biol.Chem., 300, 2024
|
|
8VCX
 
 | | Human TCR A2.13 in complex with DQ8-InsCpep | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Tran, T.M, Lim, J.J, Loh, T.J, Mannering, I.S, Rossjohn, J, Reid, H.H. | | Deposit date: | 2023-12-14 | | Release date: | 2024-08-07 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | A structural basis of T cell cross-reactivity to native and spliced self-antigens presented by HLA-DQ8.
J.Biol.Chem., 300, 2024
|
|
8VCW
 
 | | X-Ray Crystal Structure of the biotin synthase from B. obeum | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-(5-METHYL-2-OXO-IMIDAZOLIDIN-4-YL)-HEXANOIC ACID, Biotin synthase, ... | | Authors: | Lachowicz, J.C, Grove, T.L. | | Deposit date: | 2023-12-14 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Discovery of a Biotin Synthase That Utilizes an Auxiliary 4Fe-5S Cluster for Sulfur Insertion.
J.Am.Chem.Soc., 146, 2024
|
|
8VCR
 
 | | SARS-CoV-2 Spike S2 bound to Fab 54043-5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 54043-5 Fab Heavy Chain, ... | | Authors: | Johnson, N.V, McLellan, J.S. | | Deposit date: | 2023-12-14 | | Release date: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Discovery and Characterization of a Pan-betacoronavirus S2-binding antibody
To Be Published
|
|
8VCN
 
 | | GluER mutant - W66F F269Y Q293T F68Y T36E P263L | | Descriptor: | ACETATE ION, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Jeffrey, P.D, Sorigue, D.R, Liu, Y, Hyster, T.K. | | Deposit date: | 2023-12-14 | | Release date: | 2024-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Asymmetric Synthesis of alpha-Chloroamides via Photoenzymatic Hydroalkylation of Olefins.
J.Am.Chem.Soc., 146, 2024
|
|
8VCH
 
 | |
8VCD
 
 | |
8VCA
 
 | | Crystal Structure of plant Carboxylesterase 15 | | Descriptor: | Carboxylesterase 15, GLYCEROL | | Authors: | Palayam, M, Shabek, N. | | Deposit date: | 2023-12-13 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into strigolactone catabolism by carboxylesterases reveal a conserved conformational regulation.
Nat Commun, 15, 2024
|
|
8VC6
 
 | |
8VC5
 
 | |
8VC4
 
 | |
8VC3
 
 | | Voltage gated potassium ion channel Kv1.2 in complex with DTx | | Descriptor: | Kunitz-type serine protease inhibitor homolog alpha-dendrotoxin, POTASSIUM ION, Potassium voltage-gated channel subfamily A member 2 | | Authors: | Wu, Y, Sigworth, F.J. | | Deposit date: | 2023-12-13 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of Kv1.2 potassium channels, conducting and non-conducting.
Biorxiv, 2024
|
|
8VBY
 
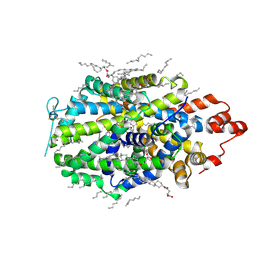 | |
8VBW
 
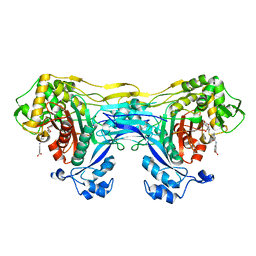 | | Structure of the monofunctional Staphylococcus aureus PBP1 in its beta-lactam (Ertapenem) inhibited form | | Descriptor: | (4R,5S)-3-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-4-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Penicillin-binding protein 1 | | Authors: | Bon, C.G, Lee, J, Caveney, N.A, Strynadka, N.C.J. | | Deposit date: | 2023-12-12 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and kinetic analysis of the monofunctional Staphylococcus aureus PBP1.
J.Struct.Biol., 216, 2024
|
|
8VBV
 
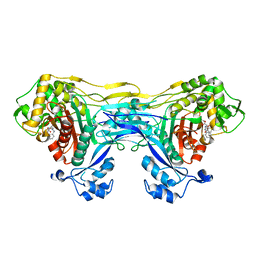 | | Structure of the monofunctional Staphylococcus aureus PBP1 in its beta-lactam (Cephalexin) inhibited form | | Descriptor: | (2S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5-methyl-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Penicillin-binding protein 1 | | Authors: | Bon, C.G, Lee, J, Caveney, N.A, Strynadka, N.C.J. | | Deposit date: | 2023-12-12 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and kinetic analysis of the monofunctional Staphylococcus aureus PBP1.
J.Struct.Biol., 216, 2024
|
|
8VBU
 
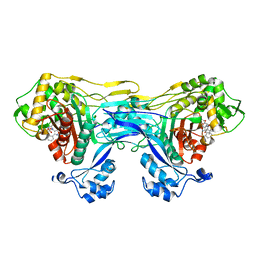 | | Structure of the monofunctional Staphylococcus aureus PBP1 in its beta-lactam (Oxacillin) inhibited form | | Descriptor: | (2R,4S)-5,5-dimethyl-2-[(1R)-1-{[(5-methyl-3-phenyl-1,2-oxazol-4-yl)carbonyl]amino}-2-oxoethyl]-1,3-thiazolidine-4-carb oxylic acid, Penicillin-binding protein 1 | | Authors: | Bon, C.G, Lee, J, Caveney, N.A, Strynadka, N.C.J. | | Deposit date: | 2023-12-12 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and kinetic analysis of the monofunctional Staphylococcus aureus PBP1.
J.Struct.Biol., 216, 2024
|
|
8VBT
 
 | |
8VBQ
 
 | | Structure of bovine anti-HIV Fab Bess7 | | Descriptor: | Bovine Fab Bess7 heavy chain, Bovine Fab Bess7 light chain | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2023-12-12 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Immunization of cows with HIV envelope trimers generates broadly neutralizing antibodies to the V2-apex from the ultra-long CDRH3 repertoire
To Be Published
|
|
8VBO
 
 | | Structure of bovine anti-HIV Fab ElsE9 | | Descriptor: | Bovine Fab ElsE9 heavy chain, Bovine Fab ElsE9 light chain, GLYCEROL, ... | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2023-12-12 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Immunization of cows with HIV envelope trimers generates broadly neutralizing antibodies to the V2-apex from the ultra-long CDRH3 repertoire
To Be Published
|
|
8VB1
 
 | | Crystal structure of HIV-1 protease with GS-9770 | | Descriptor: | (2S)-2-{(3M)-4-chloro-3-[1-(difluoromethyl)-1H-1,2,4-triazol-5-yl]phenyl}-2-[(2E,4R)-4-[4-(2-cyclopropyl-2H-1,2,3-triazol-4-yl)phenyl]-2-imino-5-oxo-4-(3,3,3-trifluoro-2,2-dimethylpropyl)imidazolidin-1-yl]ethyl [1-(difluoromethyl)cyclopropyl]carbamate, HIV-1 protease | | Authors: | Lansdon, E.B. | | Deposit date: | 2023-12-11 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Preclinical characterization of a non-peptidomimetic HIV protease inhibitor with improved metabolic stability.
Antimicrob.Agents Chemother., 68, 2024
|
|
8VAF
 
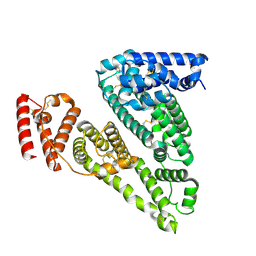 | | Cryogenic electron microscopy structure of apo human serum albumin | | Descriptor: | Serum albumin | | Authors: | Catalano, C, Lucier, K.W, To, D, Senko, S, Tran, N.L, Farwell, A.C, Silva, S.M, Dip, P.V, Poweleit, N, Scapin, G. | | Deposit date: | 2023-12-11 | | Release date: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The CryoEM structure of human serum albumin in complex with ligands.
J.Struct.Biol., 216, 2024
|
|
8VAE
 
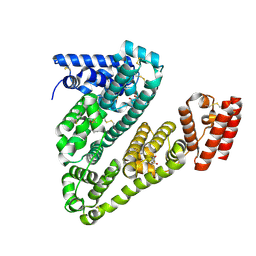 | | Cryogenic electron microscopy structure of human serum albumin in complex with salicylic acid | | Descriptor: | 2-HYDROXYBENZOIC ACID, Serum albumin | | Authors: | Catalano, C, Lucier, K.W, To, D, Senko, S, Tran, N.L, Farwell, A.C, Silva, S.M, Dip, P.V, Poweleit, N, Scapin, G. | | Deposit date: | 2023-12-11 | | Release date: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The CryoEM structure of human serum albumin in complex with ligands.
J.Struct.Biol., 216, 2024
|
|
8VAC
 
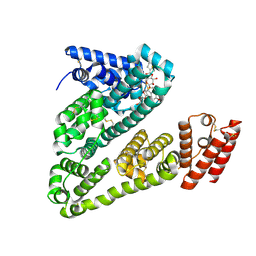 | | Cryogenic electron microscopy structure of human serum albumin in complex with teniposide | | Descriptor: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-(thiophen-2-ylmethylidene)-beta-D-glucopyranoside, Serum albumin | | Authors: | Catalano, C, Lucier, K.W, To, D, Senko, S, Tran, N.L, Farwell, A.C, Silva, S.M, Dip, P.V, Poweleit, N, Scapin, G. | | Deposit date: | 2023-12-11 | | Release date: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The CryoEM structure of human serum albumin in complex with ligands.
J.Struct.Biol., 216, 2024
|
|
