4H4X
 
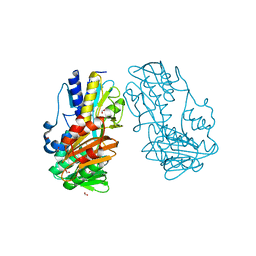 | | Crystal Structure of Ferredoxin reductase, BphA4 E175A/T176R/Q177G mutant (oxidized form) | | Descriptor: | Biphenyl dioxygenase ferredoxin reductase subunit, FLAVIN-ADENINE DINUCLEOTIDE, FORMIC ACID, ... | | Authors: | Nishizawa, A, Harada, A, Senda, M, Tachihara, Y, Muramatsu, D, Kishigami, S, Mori, S, Sugiyama, K, Senda, T, Kimura, S. | | Deposit date: | 2012-09-18 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Random Mutagenesis with the Project Assessment for Complete Conversion of Co-Factor Specificity of a Ferredoxin Reductase BphA4
To be Published
|
|
3N70
 
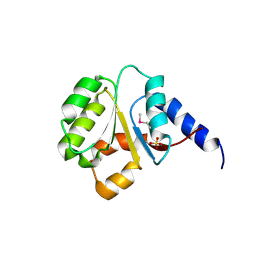 | | The Crystal Structure of the P-loop NTPase domain of the Sigma-54 transport activator from E. coli to 2.8A | | Descriptor: | SULFATE ION, Transport activator | | Authors: | Stein, A.J, Mulligan, R, Volkart, L, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-26 | | Release date: | 2010-07-21 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Crystal Structure of the P-loop NTPase domain of the Sigma-54 transport activator from E. coli to 2.8A
To be Published
|
|
4B12
 
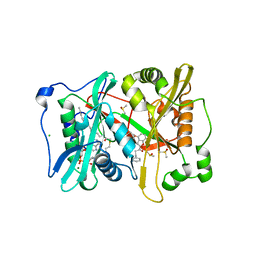 | | Plasmodium vivax N-myristoyltransferase with a bound benzofuran inhibitor (compound 23) | | Descriptor: | 1-[3-methyl-4-(piperidin-4-yloxy)-1-benzofuran-2-yl]-3-phenylpropan-1-one, 2-oxopentadecyl-CoA, CHLORIDE ION, ... | | Authors: | Yu, Z, Brannigan, J.A, Moss, D.K, Brzozowski, A.M, Wilkinson, A.J, Holder, A.A, Tate, E.W, Leatherbarrow, R.J. | | Deposit date: | 2012-07-06 | | Release date: | 2012-10-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Design and Synthesis of Inhibitors of Plasmodium Falciparum N-Myristoyltransferase, a Promising Target for Anti-Malarial Drug Discovery.
J.Med.Chem., 55, 2012
|
|
3RYF
 
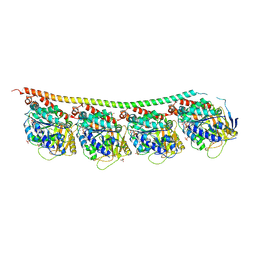 | | GTP-Tubulin: RB3 Stathmin-like domain complex | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Nawrotek, A, Knossow, M, Gigant, B. | | Deposit date: | 2011-05-11 | | Release date: | 2011-10-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | The Determinants That Govern Microtubule Assembly from the Atomic Structure of GTP-Tubulin.
J.Mol.Biol., 412, 2011
|
|
3NE7
 
 | | Crystal structure of paia n-acetyltransferase from thermoplasma acidophilum in complex with coenzyme a | | Descriptor: | ACETYLTRANSFERASE, BETA-MERCAPTOETHANOL, COENZYME A, ... | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, F.W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-08 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the novel PaiA N-acetyltransferase from Thermoplasma acidophilum involved in the negative control of sporulation and degradative enzyme production.
Proteins, 79, 2011
|
|
5YWD
 
 | |
2INW
 
 | | Crystal structure of Q83JN9 from Shigella flexneri at high resolution. Northeast Structural Genomics Consortium target SfR137. | | Descriptor: | PHOSPHATE ION, Putative structural protein | | Authors: | Kuzin, A.P, Su, M, Jayaraman, S, Vorobiev, S.M, Wang, D, Jiang, M, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-10-09 | | Release date: | 2006-10-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structures of Phd-Doc, HigA, and YeeU Establish Multiple Evolutionary Links between Microbial Growth-Regulating Toxin-Antitoxin Systems.
Structure, 18, 2010
|
|
4PVB
 
 | | Crystal structure of Aminopeptidase N in complex with the phosphonic acid analogue of leucine (D-(S)-LeuP) | | Descriptor: | Aminopeptidase N, PHOSPHATE ION, SULFATE ION, ... | | Authors: | Nocek, B, Vassiliou, S, Berlicki, L, Mulligan, R, Mucha, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-03-16 | | Release date: | 2014-06-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of Aminopeptidase N in complex with the phosphonic acid analogue of leucine (D-(S)-LeuP)
To be Published
|
|
1QQY
 
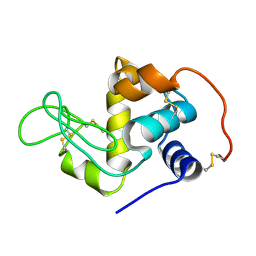 | |
1QMP
 
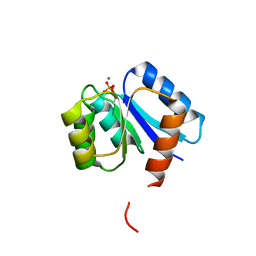 | | Phosphorylated aspartate in the crystal structure of the sporulation response regulator, Spo0A | | Descriptor: | CALCIUM ION, Stage 0 sporulation protein A | | Authors: | Lewis, R.J, Brannigan, J.A, Muchova, K, Barak, I, Wilkinson, A.J. | | Deposit date: | 1999-10-04 | | Release date: | 1999-11-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phosphorylated aspartate in the structure of a response regulator protein.
J. Mol. Biol., 294, 1999
|
|
2QO4
 
 | | Crystal structure of zebrafish liver bile acid-binding protein complexed with cholic acid | | Descriptor: | CHOLIC ACID, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Capaldi, S, Saccomani, G, Perduca, M, Monaco, H.L. | | Deposit date: | 2007-07-20 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Single Amino Acid Mutation in Zebrafish (Danio rerio) Liver Bile Acid-binding Protein Can Change the Stoichiometry of Ligand Binding.
J.Biol.Chem., 282, 2007
|
|
4DUI
 
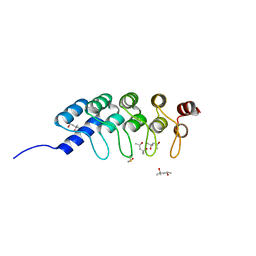 | | DARPIN D1 binding to tubulin beta chain (not in complex) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, DESIGNED ANKYRIN REPEAT PROTEIN (DARPIN) D1 | | Authors: | Pecqueur, L, Duellberg, C, Dreier, B, Wang, Q, Jiang, C, Pluckthun, A, Surrey, T, Gigant, B, Knossow, M. | | Deposit date: | 2012-02-22 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | An Anti-Tubulin Darpin Caps the Microtubule Plus-End
To be Published
|
|
2FC6
 
 | | Solution structure of the zf-CCCH domain of target of EGR1, member 1 (Nuclear) | | Descriptor: | ZINC ION, target of EGR1, member 1 | | Authors: | Dang, W, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-12 | | Release date: | 2006-06-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the zf-CCCH domain of target of EGR1, member 1 (Nuclear)
To be published
|
|
2R1D
 
 | |
3S1E
 
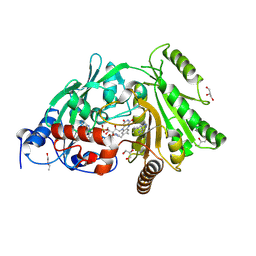 | | Pro427Gln mutant of maize cytokinin oxidase/dehydrogenase complexed with N6-isopentenyladenine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cytokinin dehydrogenase 1, ... | | Authors: | Kopecny, D, Briozzo, P, Morera, S. | | Deposit date: | 2011-05-15 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Kinetic and structural investigation of the cytokinin oxidase/dehydrogenase active site.
Febs J., 283, 2016
|
|
3O6J
 
 | | Crystal Structure of 4-Chlorocatechol Dioxygenase from Rhodococcus opacus 1CP in complex with hydroxyquinol | | Descriptor: | (2R)-3-(PHOSPHONOOXY)-2-(TETRADECANOYLOXY)PROPYL PALMITATE, CHLORIDE ION, Chlorocatechol 1,2-dioxygenase, ... | | Authors: | Ferraroni, M, Briganti, F, Kolomitseva, M, Golovleva, L. | | Deposit date: | 2010-07-29 | | Release date: | 2011-08-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-ray structures of 4-chlorocatechol 1,2-dioxygenase adducts with substituted catechols: new perspectives in the molecular basis of intradiol ring cleaving dioxygenases specificity.
J. Struct. Biol., 181, 2013
|
|
6IK5
 
 | | Crystal structure of tomato beta-galactosidase (TBG) 4 in complex with galactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-galactosidase, ... | | Authors: | Matsuyama, K, Nakae, S, Igarashi, K, Tada, T, Ishimaru, M. | | Deposit date: | 2018-10-15 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Substrate-recognition mechanism of tomato beta-galactosidase 4 using X-ray crystallography and docking simulation.
Planta, 252, 2020
|
|
4GG2
 
 | | The crystal structure of glutamate-bound human gamma-glutamyltranspeptidase 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLUTAMIC ACID, ... | | Authors: | West, M.B, Chen, Y, Wickham, S, Heroux, A, Cahill, K, Hanigan, M.H, Mooers, B.H.M. | | Deposit date: | 2012-08-04 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Novel Insights into Eukaryotic gamma-Glutamyltranspeptidase 1 from the Crystal Structure of the Glutamate-bound Human Enzyme.
J.Biol.Chem., 288, 2013
|
|
2KUP
 
 | | Solution structure of the complex of the PTB domain of SNT-2 and 19-residue peptide (aa 1571-1589) of HALK | | Descriptor: | 19-residue peptide from ALK tyrosine kinase receptor, Fibroblast growth factor receptor substrate 3 | | Authors: | Li, H, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-02-24 | | Release date: | 2010-05-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the recognition of nucleophosmin-anaplastic lymphoma kinase oncoprotein by the phosphotyrosine binding domain of Suc1-associated neurotrophic factor-induced tyrosine-phosphorylated target-2
J.Struct.Funct.Genom., 11, 2010
|
|
2WPN
 
 | | Structure of the oxidised, as-isolated NiFeSe hydrogenase from D. vulgaris Hildenborough | | Descriptor: | 3-[DODECYL(DIMETHYL)AMMONIO]PROPANE-1-SULFONATE, BIS-(MU-2-OXO),[(MU-3--SULFIDO)-BIS(MU-2--SULFIDO)-TRIS(CYS-S)-TRI-IRON] (AQUA)(GLU-O)IRON(II), CARBONMONOXIDE-(DICYANO) IRON, ... | | Authors: | Marques, M.C, Coelho, R, De Lacey, A.L, Pereira, I.A.C, Matias, P.M. | | Deposit date: | 2009-08-07 | | Release date: | 2010-01-12 | | Last modified: | 2019-07-10 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The three-dimensional structure of [NiFeSe] hydrogenase from Desulfovibrio vulgaris Hildenborough: a hydrogenase without a bridging ligand in the active site in its oxidised, "as-isolated" state.
J.Mol.Biol., 396, 2010
|
|
7EOK
 
 | | Crystal structure of the Pepper aptamer in complex with HBC485 | | Descriptor: | 4-[(~{Z})-1-cyano-2-[5-[2-(dimethylamino)ethyl-methyl-amino]pyrazin-2-yl]ethenyl]benzenecarbonitrile, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Huang, K.Y, Ren, A.M. | | Deposit date: | 2021-04-22 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-based investigation of fluorogenic Pepper aptamer.
Nat.Chem.Biol., 17, 2021
|
|
7EOO
 
 | | Crystal structure of the Pepper aptamer in complex with HBC525 | | Descriptor: | (~{E})-2-(1,3-benzoxazol-2-yl)-3-[4-[2-hydroxyethyl(methyl)amino]phenyl]prop-2-enenitrile, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Huang, K.Y, Ren, A.M. | | Deposit date: | 2021-04-22 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structure-based investigation of fluorogenic Pepper aptamer.
Nat.Chem.Biol., 17, 2021
|
|
7F0L
 
 | | STRUCTURE OF PHOTOSYNTHETIC LH1-RC SUPER-COMPLEX OF RHODOBACTER SPHAEROIDES MONOMER | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, Antenna pigment protein beta chain, BACTERIOCHLOROPHYLL A, ... | | Authors: | Tani, K, Nagashima, V.P, Kanno, R, Kawamura, S, Kikuchi, R, Ji, X.-C, Hall, M, Yu, L.-J, Kimura, Y, Madigan, M.T, Mizoguchi, A, Humbel, B.M, Wang-Otomo, Z.-Y. | | Deposit date: | 2021-06-05 | | Release date: | 2021-11-10 | | Last modified: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | A previously unrecognized membrane protein in the Rhodobacter sphaeroides LH1-RC photocomplex.
Nat Commun, 12, 2021
|
|
7EOP
 
 | | Crystal structure of the Pepper aptamer in complex with HBC620 | | Descriptor: | 4-[(~{Z})-1-cyano-2-[5-[2-hydroxyethyl(methyl)amino]thieno[3,2-b]thiophen-2-yl]ethenyl]benzenecarbonitrile, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Huang, K.Y, Ren, A.M. | | Deposit date: | 2021-04-22 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based investigation of fluorogenic Pepper aptamer.
Nat.Chem.Biol., 17, 2021
|
|
7EOL
 
 | | Crystal structure of the Pepper aptamer in complex with HBC497 | | Descriptor: | 4-[(~{Z})-1-cyano-2-[5-[2-hydroxyethyl(methyl)amino]pyrazin-2-yl]ethenyl]benzenecarbonitrile, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Huang, K.Y, Ren, A.M. | | Deposit date: | 2021-04-22 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.309 Å) | | Cite: | Structure-based investigation of fluorogenic Pepper aptamer.
Nat.Chem.Biol., 17, 2021
|
|
