8K0M
 
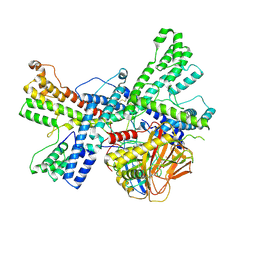 | | Human collagen prolyl processing enzyme complex, P3H1/CRTAP/PPIB heterotrimer, bound to 2-oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Cartilage-associated protein, ... | | Authors: | Li, W, Peng, J, Yao, D, Rao, B, Xia, Y, Wang, Q, Li, S, Cao, M, Shen, Y, Ma, P, Liao, R, Qin, A, Zhao, J, Cao, Y. | | Deposit date: | 2023-07-09 | | Release date: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | The structural basis for the collagen processing by human P3H1/CRTAP/PPIB ternary complex.
Nat Commun, 15, 2024
|
|
8K0F
 
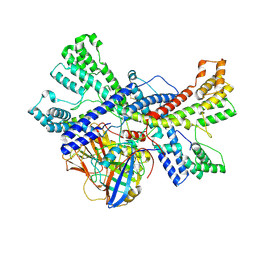 | | Human collagen prolyl processing enzyme complex, P3H1/CRTAP/PPIB heterotrimer, in its apo state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cartilage-associated protein, FE (III) ION, ... | | Authors: | Li, W, Peng, J, Yao, D, Rao, B, Xia, Y, Wang, Q, Li, S, Cao, M, Shen, Y, Ma, P, Liao, R, Qin, A, Zhao, J, Cao, Y. | | Deposit date: | 2023-07-08 | | Release date: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | The structural basis for the collagen processing by human P3H1/CRTAP/PPIB ternary complex.
Nat Commun, 15, 2024
|
|
8K17
 
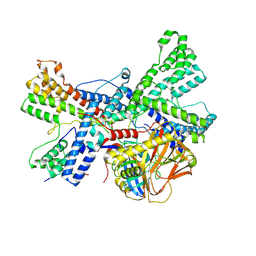 | | Human collagen prolyl processing enzyme complex, P3H1/CRTAP/PPIB heterotrimer, bound to collagen alpha-1(I) chain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cartilage-associated protein, FE (III) ION, ... | | Authors: | Li, W, Peng, J, Yao, D, Rao, B, Xia, Y, Wang, Q, Li, S, Cao, M, Shen, Y, Ma, P, Liao, R, Qin, A, Zhao, J, Cao, Y. | | Deposit date: | 2023-07-10 | | Release date: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | The structural basis for the collagen processing by human P3H1/CRTAP/PPIB ternary complex.
Nat Commun, 15, 2024
|
|
7JV7
 
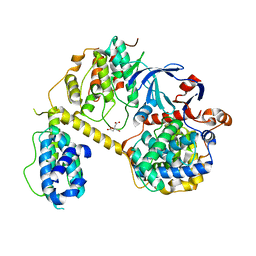 | | Crystal Structure of the yeast RNA Pol II CTD kinase CTDK-1 complex | | Descriptor: | CITRATE ANION, CTD kinase subunit alpha, CTD kinase subunit beta, ... | | Authors: | Xie, Y, Ren, Y. | | Deposit date: | 2020-08-20 | | Release date: | 2021-01-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.850553 Å) | | Cite: | Structure and activation mechanism of the yeast RNA Pol II CTD kinase CTDK-1 complex.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1TBK
 
 | |
1LRA
 
 | | CRYSTALLOGRAPHIC STUDY OF GLU 58 ALA RNASE T1(ASTERISK)2'-GUANOSINE MONOPHOSPHATE AT 1.9 ANGSTROMS RESOLUTION | | Descriptor: | GUANOSINE-2'-MONOPHOSPHATE, RIBONUCLEASE T1, SODIUM ION | | Authors: | Pletinckx, J, Steyaert, J, Choe, H.-W, Heinemann, U, Wyns, L. | | Deposit date: | 1993-10-01 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic study of Glu58Ala RNase T1 x 2'-guanosine monophosphate at 1.9-A resolution.
Biochemistry, 33, 1994
|
|
2M6O
 
 | | The actinobacterial transcription factor RbpA binds to the principal sigma subunit of RNA polymerase | | Descriptor: | Uncharacterized protein | | Authors: | Liu, B, Tabib-Salazar, A, Doughty, P, Lewis, R, Ghosh, S, Parsy, M, Simpson, P, Matthews, S, Paget, M. | | Deposit date: | 2013-04-06 | | Release date: | 2013-05-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The actinobacterial transcription factor RbpA binds to the principal sigma subunit of RNA polymerase.
Nucleic Acids Res., 41, 2013
|
|
2M6P
 
 | |
4U96
 
 | |
4DXX
 
 | | TGT K52M mutant crystallized at pH 8.5 | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Jakobi, S, Ritschel, T, Heine, A, Klebe, G. | | Deposit date: | 2012-02-28 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Hot-spot analysis to dissect the functional protein-protein interface of a tRNA-modifying enzyme.
Proteins, 82, 2014
|
|
2JH9
 
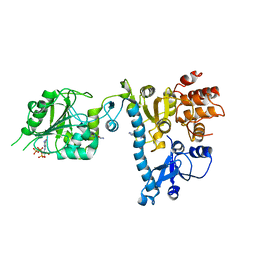 | | The structure of bluetongue virus VP4 reveals a multifunctional RNA- capping production-line | | Descriptor: | GUANINE, GUANOSINE-5'-TRIPHOSPHATE, VP4 CORE PROTEIN | | Authors: | Sutton, G, Grimes, J.M, Stuart, D.I, Roy, P. | | Deposit date: | 2007-02-21 | | Release date: | 2007-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Bluetongue Virus Vp4 is an RNA-Capping Assembly Line.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2JHP
 
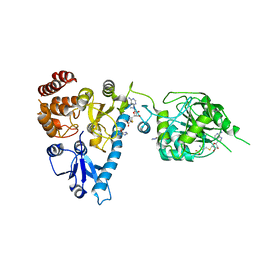 | | The structure of bluetongue virus VP4 reveals a multifunctional RNA- capping production-line | | Descriptor: | GUANINE, S-ADENOSYL-L-HOMOCYSTEINE, VP4 CORE PROTEIN | | Authors: | Sutton, G, Grimes, J.M, Stuart, D.I, Roy, P. | | Deposit date: | 2007-02-23 | | Release date: | 2007-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Bluetongue Virus Vp4 is an RNA-Capping Assembly Line.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2JHC
 
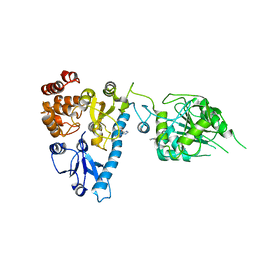 | | The structure of bluetongue virus VP4 reveals a multifunctional RNA- capping production-line | | Descriptor: | GUANINE, VP4 CORE PROTEIN | | Authors: | Sutton, G, Grimes, J.M, Stuart, D.I, Roy, P. | | Deposit date: | 2007-02-21 | | Release date: | 2007-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Bluetongue Virus Vp4 is an RNA-Capping Assembly Line.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2L5D
 
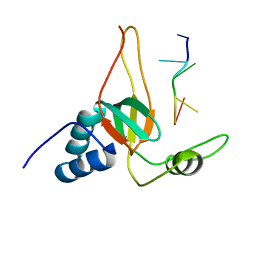 | | Solution Structures of human PIWI-like 1 PAZ domain with ssRNA (5'-pUGACA) | | Descriptor: | 5'-R(*UP*GP*AP*CP*A)-3', Piwi-like protein 1 | | Authors: | Zeng, L, Zhang, Q, Yan, K, Zhou, M. | | Deposit date: | 2010-10-29 | | Release date: | 2011-04-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural insights into piRNA recognition by the human PIWI-like 1 PAZ domain.
Proteins, 79, 2011
|
|
2JHA
 
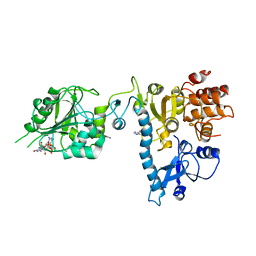 | | The structure of bluetongue virus VP4 reveals a multifunctional RNA- capping production-line | | Descriptor: | DIGUANOSINE-5'-TRIPHOSPHATE, GUANINE, VP4 CORE PROTEIN | | Authors: | Sutton, G, Grimes, J.M, Stuart, D.I, Roy, P. | | Deposit date: | 2007-02-21 | | Release date: | 2007-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Bluetongue Virus Vp4 is an RNA-Capping Assembly Line.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2BCW
 
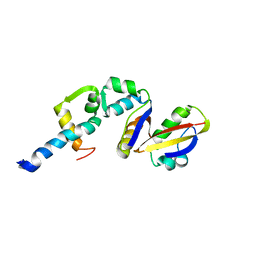 | | Coordinates of the N-terminal domain of ribosomal protein L11,C-terminal domain of ribosomal protein L7/L12 and a portion of the G' domain of elongation factor G, as fitted into cryo-em map of an Escherichia coli 70S*EF-G*GDP*fusidic acid complex | | Descriptor: | 50S ribosomal protein L11, 50S ribosomal protein L7/L12, Elongation factor G | | Authors: | Datta, P.P, Sharma, M.R, Qi, L, Frank, J, Agrawal, R.K. | | Deposit date: | 2005-10-19 | | Release date: | 2005-12-20 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (11.2 Å) | | Cite: | Interaction of the G' Domain of Elongation Factor G and the C-Terminal Domain of Ribosomal Protein L7/L12 during Translocation as Revealed by Cryo-EM.
Mol.Cell, 20, 2005
|
|
4MEM
 
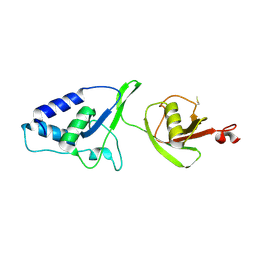 | | Crystal Structure of the rat USP11 DUSP-UBL domains | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 11 | | Authors: | Harper, S, Gratton, H.E, Cornaciu, I, Oberer, M, Scott, D.J, Emsley, J, Dreveny, I. | | Deposit date: | 2013-08-27 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structure and Catalytic Regulatory Function of Ubiquitin Specific Protease 11 N-Terminal and Ubiquitin-like Domains.
Biochemistry, 53, 2014
|
|
4MEL
 
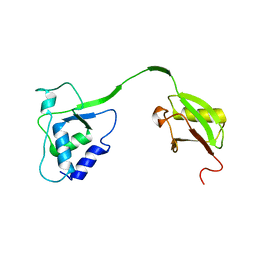 | | Crystal Structure of the human USP11 DUSP-UBL domains | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 11 | | Authors: | Harper, S, Gratton, H.E, Cornaciu, I, Oberer, M, Scott, D.J, Emsley, J, Dreveny, I. | | Deposit date: | 2013-08-27 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.899 Å) | | Cite: | Structure and Catalytic Regulatory Function of Ubiquitin Specific Protease 11 N-Terminal and Ubiquitin-like Domains.
Biochemistry, 53, 2014
|
|
4IQZ
 
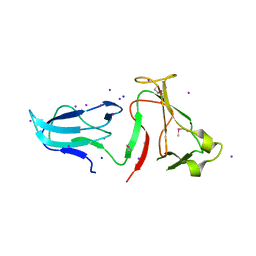 | | The crystal structure of a large insert in RNA polymerase (RpoC) subunit from E. coli | | Descriptor: | DNA-directed RNA polymerase subunit beta', IODIDE ION, SODIUM ION | | Authors: | Bhandari, V, Sugiman-Marangos, S.N, Naushad, H.S, Gupta, R.S, Junop, M.S. | | Deposit date: | 2013-01-14 | | Release date: | 2013-02-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of a large insert in RNA polymerase (RpoC) subunit from E. coli
To be Published
|
|
1O1W
 
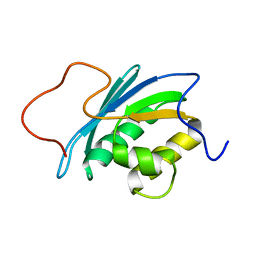 | | SOLUTION STRUCTURE OF THE RNASE H DOMAIN OF THE HIV-1 REVERSE TRANSCRIPTASE IN THE PRESENCE OF MAGNESIUM | | Descriptor: | RIBONUCLEASE H | | Authors: | Pari, K, Mueller, G.A, Derose, E.F, Kirby, T.W, London, R.E. | | Deposit date: | 2003-02-12 | | Release date: | 2003-02-18 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Rnase H Domain of the HIV-1 Reverse Transcriptase in the Presence of Magnesium
Biochemistry, 42, 2003
|
|
5DHB
 
 | | Cooperativity and Downstream Binding in RNA Replication | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, RNA (5'-R(*(LCC)P*(TLN)P*(LCG)P*UP*AP*CP*A)-3') | | Authors: | Zhang, W, Fahrenbach, A.C, Tam, C.P, Szostak, J.W. | | Deposit date: | 2015-08-30 | | Release date: | 2016-12-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Unusual Base-Pairing Interactions in Monomer-Template Complexes.
ACS Cent Sci, 2, 2016
|
|
5DHC
 
 | | Cooperativity and Downstream Binding in RNA Replication | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, MAGNESIUM ION, RNA (5'-R(*(LCC)P*(LCC)P*(LCA)P*(LCG)P*AP*CP*UP*UP*AP*AP*GP*UP*CP*U)-3') | | Authors: | Zhang, W, Fahrenbach, A.C, Tam, C.P, Szostak, J.W. | | Deposit date: | 2015-08-30 | | Release date: | 2016-12-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Unusual Base-Pairing Interactions in Monomer-Template Complexes.
ACS Cent Sci, 2, 2016
|
|
2L0I
 
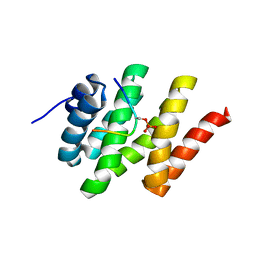 | | Solution structure of Rtt103 CTD-interacting domain bound to a Ser2 phosphorylated CTD peptide | | Descriptor: | DNA-directed RNA polymerase, Regulator of Ty1 transposition protein 103 | | Authors: | Lunde, B.M, Reichow, S.L, Kim, M, Suh, H, Leeper, T.C, Yang, F, Mutschler, H, Buratowski, S, Meinhart, A, Varani, G. | | Deposit date: | 2010-07-06 | | Release date: | 2010-09-08 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Cooperative interaction of transcription termination factors with the RNA polymerase II C-terminal domain.
Nat.Struct.Mol.Biol., 17, 2010
|
|
1MI6
 
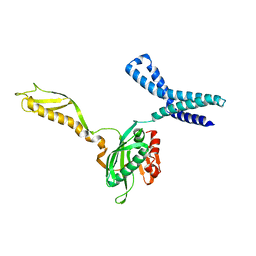 | | Docking of the modified RF2 X-ray structure into the Low Resolution Cryo-EM map of RF2 E.coli 70S Ribosome | | Descriptor: | peptide chain release factor RF-2 | | Authors: | Rawat, U.B.S, Zavialov, A.V, Sengupta, J, Valle, M, Grassucci, R.A, Linde, J, Vestergaard, B, Ehrenberg, M, Frank, J. | | Deposit date: | 2002-08-22 | | Release date: | 2003-01-14 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (12.8 Å) | | Cite: | A cryo-electron microscopic study of ribosome-bound termination factor RF2
Nature, 421, 2003
|
|
7KCJ
 
 | |
