1WU3
 
 | |
1QJ3
 
 | | Crystal structure of 7,8-diaminopelargonic acid synthase in complex with 7-keto-8-aminopelargonic acid | | Descriptor: | 7,8-DIAMINOPELARGONIC ACID SYNTHASE, 7-KETO-8-AMINOPELARGONIC ACID, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Kaeck, H, Sandmark, J, Gibson, K.J, Lindqvist, Y, Schneider, G. | | Deposit date: | 1999-06-21 | | Release date: | 2000-06-22 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Diaminopelargonic Acid Synthase; Evolutionary Relationships between Pyridoxal-5'-Phosphate Dependent Enzymes
J.Mol.Biol., 291, 1999
|
|
1XNE
 
 | | Solution Structure of Pyrococcus furiosus Protein PF0470: The Northeast Structural Genomics Consortium Target PfR14 | | Descriptor: | hypothetical protein PF0469 | | Authors: | Liu, G, Xiao, R, Parish, D, Ma, L, Sukumaran, D, Acton, T, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-10-04 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR data collection and analysis protocol for high-throughput protein structure determination.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1XN6
 
 | | Solution Structure of Northeast Structural Genomics Target Protein BcR68 encoded in gene Q816V6 of B. cereus | | Descriptor: | hypothetical protein BC4709 | | Authors: | Liu, G, Acton, T, Parish, D, Ma, L, Xu, D, Xiao, R, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-10-04 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR data collection and analysis protocol for high-throughput protein structure determination.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1R4W
 
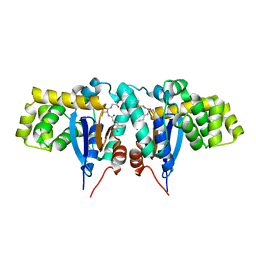 | | Crystal structure of Mitochondrial class kappa glutathione transferase | | Descriptor: | GLUTATHIONE, Glutathione S-transferase, mitochondrial | | Authors: | Ladner, J.E, Parsons, J.F, Rife, C.L, Gilliland, G.L, Armstrong, R.N. | | Deposit date: | 2003-10-08 | | Release date: | 2004-02-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Parallel Evolutionary Pathways for Glutathione Transferases: Structure and Mechanism of the Mitochondrial Class Kappa Enzyme rGSTK1-1
Biochemistry, 43, 2004
|
|
1S6Q
 
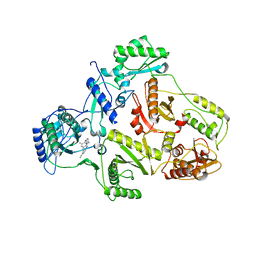 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH JANSSEN-R147681 | | Descriptor: | 4-[4-(2,4,6-TRIMETHYL-PHENYLAMINO)-PYRIMIDIN-2-YLAMINO]-BENZONITRILE, POL polyprotein [Contains: Reverse transcriptase] | | Authors: | Das, K, Arnold, E. | | Deposit date: | 2004-01-26 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Roles of Conformational and Positional Adaptability in Structure-Based Design of TMC125-R165335 (Etravirine) and Related Non-nucleoside Reverse Transcriptase Inhibitors That Are Highly Potent and Effective against Wild-Type and Drug-Resistant HIV-1 Variants.
J.Med.Chem., 47, 2004
|
|
1S9E
 
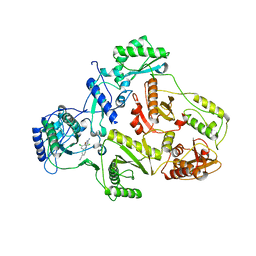 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH JANSSEN-R129385 | | Descriptor: | 4-[4-AMINO-6-(2,6-DICHLORO-PHENOXY)-[1,3,5]TRIAZIN-2-YLAMINO]-BENZONITRILE, POL polyprotein [Contains: Reverse transcriptase], POL polyprotein [Contains:Reverse transcriptase] | | Authors: | Das, K, Clark Jr, A.D, Ludovici, D.W, Kukla, M.J, Decorte, B, Lewi, P.J, Hughes, S.H, Janssen, P.A, Arnold, E. | | Deposit date: | 2004-02-04 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Roles of Conformational and Positional Adaptability in Structure-Based Design of TMC125-R165335 (Etravirine) and Related Non-nucleoside Reverse Transcriptase Inhibitors That Are Highly Potent and Effective against Wild-Type and Drug-Resistant HIV-1 Variants.
J.Med.Chem., 47, 2004
|
|
1S9G
 
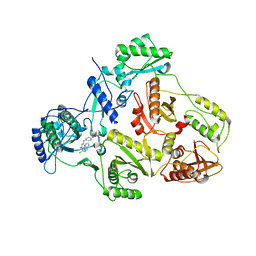 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH JANSSEN-R120394. | | Descriptor: | 4-[4-AMINO-6-(5-CHLORO-1H-INDOL-4-YLMETHYL)-[1,3,5]TRIAZIN-2-YLAMINO]-BENZONITRILE, POL polyprotein [Contains: Reverse transcriptase] | | Authors: | Das, K, Clark Jr, A.D, Ludovici, D.W, Kukla, M.J, Decorte, B, Lewi, P.J, Hughes, S.H, Janssen, P.A, Arnold, E. | | Deposit date: | 2004-02-04 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Roles of Conformational and Positional Adaptability in Structure-Based Design of TMC125-R165335 (Etravirine) and Related Non-nucleoside Reverse Transcriptase Inhibitors That Are Highly Potent and Effective against Wild-Type and Drug-Resistant HIV-1 Variants.
J.Med.Chem., 47, 2004
|
|
1SQF
 
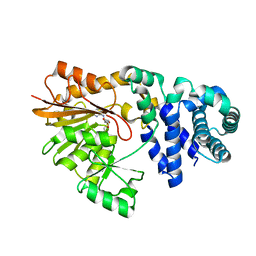 | | The crystal structure of E. coli Fmu binary complex with S-Adenosylmethionine at 2.1 A resolution | | Descriptor: | S-ADENOSYLMETHIONINE, SUN protein | | Authors: | Foster, P.G, Nunes, C.R, Greene, P, Moustakas, D, Stroud, R.M. | | Deposit date: | 2004-03-18 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The First Structure of an RNA m5C Methyltransferase,
Fmu, Provides Insight into Catalytic Mechanism
and Specific Binding of RNA Substrate
Structure, 11, 2003
|
|
1YP7
 
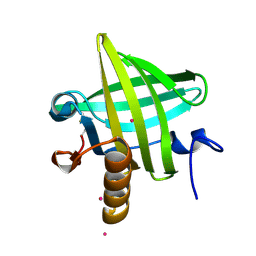 | | Van der Waals Interactions Dominate Hydrophobic Association in a Protein Binding Site Occluded From Solvent Water | | Descriptor: | CADMIUM ION, MAJOR URINARY PROTEIN 1 | | Authors: | Barratt, E, Bingham, R.J, Warner, D.J, Laughton, C.A, Phillips, S.E.V, Homans, S.W. | | Deposit date: | 2005-01-30 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Van der Waals Interactions Dominate Ligand-Protein Association in a Protein Binding Site Occluded from Solvent Water
J.Am.Chem.Soc., 127, 2005
|
|
1SUQ
 
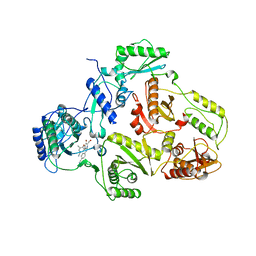 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH JANSSEN-R185545 | | Descriptor: | (6-[4-(AMINOMETHYL)-2,6-DIMETHYLPHENOXY]-2-{[4-(AMINOMETHYL)PHENYL]AMINO}-5-BROMOPYRIMIDIN-4-YL)METHANOL, MAGNESIUM ION, REVERSE TRANSCRIPTASE | | Authors: | Das, K, Arnold, E. | | Deposit date: | 2004-03-26 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Roles of Conformational and Positional Adaptability in Structure-Based Design of TMC125-R165335 (Etravirine) and Related Non-nucleoside Reverse Transcriptase Inhibitors That Are Highly Potent and Effective against Wild-Type and Drug-Resistant HIV-1 Variants
J.Med.Chem., 47, 2004
|
|
1YP6
 
 | | Van der Waals Interactions Dominate Hydrophobic Association in a Protein Binding Site Occluded From Solvent Water | | Descriptor: | 2-ISOBUTYL-3-METHOXYPYRAZINE, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Barratt, E, Bingham, R.J, Warner, D.J, Laughton, C.A, Phillips, S.E.V, Homans, S.W. | | Deposit date: | 2005-01-30 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Van der Waals Interactions Dominate Ligand-Protein Association in a Protein Binding Site Occluded from Solvent Water
J.Am.Chem.Soc., 127, 2005
|
|
1XN8
 
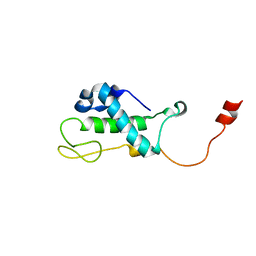 | | Solution Structure of Bacillus subtilis Protein yqbG: The Northeast Structural Genomics Consortium Target SR215 | | Descriptor: | Hypothetical protein yqbG | | Authors: | Liu, G, Ma, L, Shen, Y, Acton, T, Atreya, H.S, Xiao, R, Joachimiak, A, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-10-04 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR data collection and analysis protocol for high-throughput protein structure determination.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1VC8
 
 | |
1XBD
 
 | | INTERNAL XYLAN BINDING DOMAIN FROM CELLULOMONAS FIMI XYLANASE D, NMR, 5 STRUCTURES | | Descriptor: | XYLANASE D | | Authors: | Simpson, P.J, Bolam, D.N, Cooper, A, Ciruela, A, Hazlewood, G.P, Gilbert, H.J, Williamson, M.P. | | Deposit date: | 1998-10-16 | | Release date: | 1999-07-21 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | A family IIb xylan-binding domain has a similar secondary structure to a homologous family IIa cellulose-binding domain but different ligand specificity.
Structure Fold.Des., 7, 1999
|
|
1SV5
 
 | | CRYSTAL STRUCTURE OF K103N MUTANT HIV-1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH JANSSEN-R165335 | | Descriptor: | 4-({6-AMINO-5-BROMO-2-[(4-CYANOPHENYL)AMINO]PYRIMIDIN-4-YL}OXY)-3,5-DIMETHYLBENZONITRILE, Reverse Transcriptase | | Authors: | Das, K, Arnold, E. | | Deposit date: | 2004-03-27 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Roles of Conformational and Positional Adaptability in Structure-Based Design of TMC125-R165335 (Etravirine) and Related Non-nucleoside Reverse Transcriptase Inhibitors That Are Highly Potent and Effective against Wild-Type and Drug-Resistant HIV-1 Variants
J.Med.Chem., 47, 2004
|
|
1Z00
 
 | | Solution structure of the C-terminal domain of ERCC1 complexed with the C-terminal domain of XPF | | Descriptor: | DNA excision repair protein ERCC-1, DNA repair endonuclease XPF | | Authors: | Tripsianes, K, Folkers, G, Ab, E, Das, D, Odijk, H, Jaspers, N.G.J, Hoeijmakers, J.H.J, Kaptein, R, Boelens, R. | | Deposit date: | 2005-03-01 | | Release date: | 2005-12-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The Structure of the Human ERCC1/XPF Interaction Domains Reveals a Complementary Role for the Two Proteins in Nucleotide Excision Repair
Structure, 13, 2005
|
|
1VCD
 
 | | Crystal Structure of a T.thermophilus HB8 Ap6A hydrolase Ndx1 | | Descriptor: | GLYCEROL, Ndx1, SULFATE ION | | Authors: | Iwai, T, Nakagawa, N, Kuramitsu, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-03-05 | | Release date: | 2005-04-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Nudix Protein Ndx1 from Thermus thermophilus HB8 in binary complex with diadenosine hexaphosphate
To be Published
|
|
1VC9
 
 | | Crystal Structure of a T.thermophilus HB8 Ap6A hydrolase E50Q mutant-Mg2+-ATP complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Ndx1 | | Authors: | Iwai, T, Nakagawa, N, Kuramitsu, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-03-05 | | Release date: | 2005-04-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Nudix Protein Ndx1 from Thermus thermophilus HB8 in binary complex with diadenosine hexaphosphate
To be Published
|
|
1ZO9
 
 | | Crystal Structure Of The Wild Type Heme Domain Of P450BM-3 with N-palmitoylmethionine | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Bifunctional P-450:NADPH-P450 reductase, GLYCEROL, ... | | Authors: | Hegda, A, Chen, B, Tomchick, D.R, Bondlela, M, Haines, D.C, Schaffer, N, Machius, M, Graham, S.E, Peterson, J.A. | | Deposit date: | 2005-05-12 | | Release date: | 2006-08-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Interactions of substrates at the surface of P450s can greatly enhance substrate potency.
Biochemistry, 46, 2007
|
|
1ZP5
 
 | | Crystal structure of the complex between MMP-8 and a N-hydroxyurea inhibitor | | Descriptor: | CALCIUM ION, N-{2-[(4'-CYANO-1,1'-BIPHENYL-4-YL)OXY]ETHYL}-N'-HYDROXY-N-METHYLUREA, Neutrophil collagenase, ... | | Authors: | Campestre, C, Agamennone, M, Tortorella, P, Preziuso, S, Biasone, A, Gavuzzo, E, Pochetti, G, Mazza, F, Tschesche, H, Gallina, C. | | Deposit date: | 2005-05-16 | | Release date: | 2005-12-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | N-Hydroxyurea as zinc binding group in matrix metalloproteinase inhibition: Mode of binding in a complex with MMP-8.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
1Z4R
 
 | | Human GCN5 Acetyltransferase | | Descriptor: | ACETYL COENZYME *A, General control of amino acid synthesis protein 5-like 2 | | Authors: | Dong, A, Bernstein, G, Schuetz, A, Antoshenko, T, Wu, H, Loppnau, P, Sundstrom, M, Arrowsmith, C, Edwards, A, Bochkarev, A, Plotnikov, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-03-16 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of a binary complex between human GCN5 histone acetyltransferase domain and acetyl coenzyme A
Proteins, 68, 2007
|
|
1ZAK
 
 | |
1BTU
 
 | | PORCINE PANCREATIC ELASTASE COMPLEXED WITH (3S, 4R)-1-TOLUENESULPHONYL-3-ETHYL-AZETIDIN-2-ONE-4-CARBOXYLIC ACID | | Descriptor: | (3R)-3-ethyl-N-[(4-methylphenyl)sulfonyl]-L-aspartic acid, CALCIUM ION, ELASTASE, ... | | Authors: | Wilmouth, R.C, Clifton, I.J, Schofield, C.J. | | Deposit date: | 1998-09-01 | | Release date: | 1999-02-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Inhibition of elastase by N-sulfonylaryl beta-lactams: anatomy of a stable acyl-enzyme complex.
Biochemistry, 37, 1998
|
|
1PMD
 
 | | PENICILLIN-BINDING PROTEIN 2X (PBP-2X) | | Descriptor: | PEPTIDOGLYCAN SYNTHESIS MULTIFUNCTIONAL ENZYME | | Authors: | Pares, S, Mouz, N, Dideberg, O. | | Deposit date: | 1996-02-05 | | Release date: | 1997-02-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | X-ray structure of Streptococcus pneumoniae PBP2x, a primary penicillin target enzyme.
Nat.Struct.Biol., 3, 1996
|
|
