2K73
 
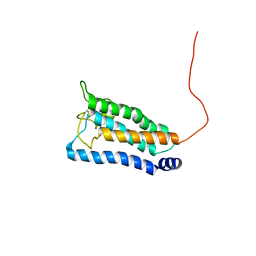 | | Solution NMR structure of integral membrane protein DsbB | | Descriptor: | Disulfide bond formation protein B | | Authors: | Zhou, Y, Cierpicki, T, Flores Jimenez, R.H, Lukasik, S.M, Ellena, J.F, Cafiso, D.S, Kadokura, H, Beckwith, J, Bushweller, J.H. | | Deposit date: | 2008-08-01 | | Release date: | 2008-10-07 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the integral membrane enzyme DsbB: functional insights into DsbB-catalyzed disulfide bond formation.
Mol.Cell, 31, 2008
|
|
1GT0
 
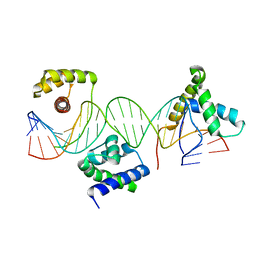 | | Crystal structure of a POU/HMG/DNA ternary complex | | Descriptor: | 5'-D(*AP*TP*CP*CP*CP*AP*TP*TP*AP*GP* CP*AP*TP*CP*CP*AP*AP*AP*CP*AP*AP*AP*GP*A)-3', 5'-D(*TP*TP*CP*TP*TP*TP*GP*TP*TP*TP* GP*GP*AP* TP*GP*CP*TP*AP*AP*TP*GP*GP*GP*A)-3', OCTAMER-BINDING TRANSCRIPTION FACTOR 1, ... | | Authors: | Remenyi, A, Wilmanns, M. | | Deposit date: | 2002-01-09 | | Release date: | 2003-01-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of a POU/Hmg/DNA Ternary Complex Suggests Differential Assembly of Oct4 and Sox2 on Two Enhancers
Genes Dev., 17, 2003
|
|
2KNH
 
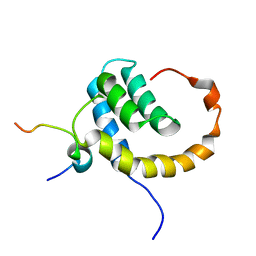 | | The Solution structure of the eTAFH domain of AML1-ETO complexed with HEB peptide | | Descriptor: | Protein CBFA2T1, Transcription factor 12 | | Authors: | Park, S, Cierpicki, T, Tonelli, M, Bushweller, J.H. | | Deposit date: | 2009-08-25 | | Release date: | 2009-10-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the AML1-ETO eTAFH domain-HEB peptide complex and its contribution to AML1-ETO activity.
Blood, 113, 2009
|
|
1GRF
 
 | |
2KDQ
 
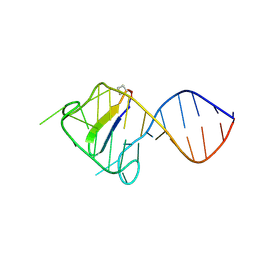 | |
1GVY
 
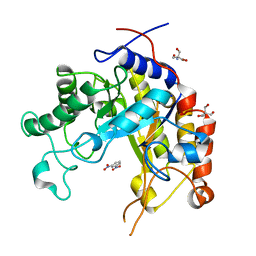 | | Substrate distorsion by beta-mannanase from Pseudomonas cellulosa | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DINITROPHENYLENE, MANNAN ENDO-1,4-BETA-MANNOSIDASE, ... | | Authors: | Ducros, V, Zechel, D.L, Gilbert, H.J, Szabo, L, Withers, S.G, Davies, G.J. | | Deposit date: | 2002-02-28 | | Release date: | 2002-09-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Substrate Distortion by a Beta-Mannanase: Snapshots of the Michaelis and Covalent-Intermediate Complexes Suggest a B2,5 Conformation for the Transition State
Angew.Chem.Int.Ed.Engl., 41, 2002
|
|
2KDW
 
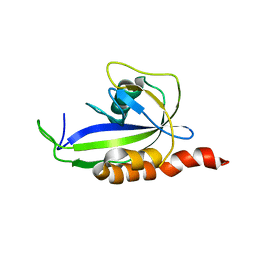 | |
1GRJ
 
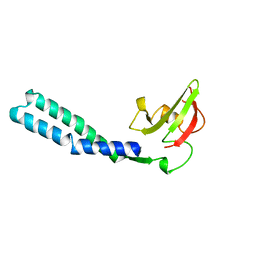 | |
1H10
 
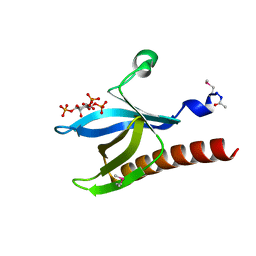 | | HIGH RESOLUTION STRUCTURE OF THE PLECKSTRIN HOMOLOGY DOMAIN OF PROTEIN KINASE B/AKT BOUND TO INS(1,3,4,5)-TETRAKISPHOPHATE | | Descriptor: | INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, RAC-ALPHA SERINE/THREONINE KINASE | | Authors: | Thomas, C.C, Deak, M, Alessi, D.R, Van Aalten, D.M.F. | | Deposit date: | 2002-07-01 | | Release date: | 2003-06-27 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High Resolution Structure of the Pleckstrin Homology Domain of Protein Kinase B/Akt Bound to Phosphatidylinositol (3,4,5)-Trisphosphate
Curr.Biol., 12, 2002
|
|
2KHR
 
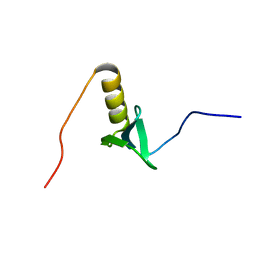 | |
1H32
 
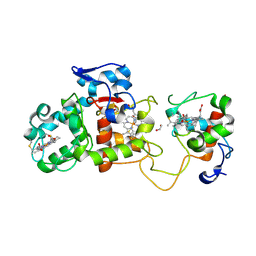 | | Reduced SoxAX complex from Rhodovulum sulfidophilum | | Descriptor: | 1,2-ETHANEDIOL, CYTOCHROME C, DIHEME CYTOCHROME C, ... | | Authors: | Bamford, V.A, Bruno, S, Rasmussen, T, Appia-Ayme, C, Cheesman, M.R, Berks, B.C, Hemmings, A.M. | | Deposit date: | 2002-08-21 | | Release date: | 2002-11-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for the Oxidation of Thiosulfate by a Sulfur Cycle Enzyme
Embo J., 21, 2002
|
|
1H9Z
 
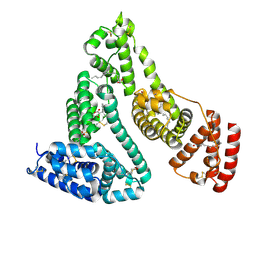 | |
2KN8
 
 | | NMR structure of the C-terminal domain of pUL89 | | Descriptor: | DNA cleavage and packaging protein large subunit, UL89 | | Authors: | Couvreux, A, Hantz, S, Marquant, R, Champier, G, Alain, S, Morellet, N, Bouaziz, S. | | Deposit date: | 2009-08-18 | | Release date: | 2010-06-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Insight into the structure of the pUL89 C-terminal domain of the human cytomegalovirus terminase complex.
Proteins, 78, 2010
|
|
1H12
 
 | | Structure of a cold-adapted family 8 xylanase | | Descriptor: | ENDO-1,4-BETA-XYLANASE, alpha-D-xylopyranose, beta-D-xylopyranose | | Authors: | Van Petegem, F, Collins, T, Meuwis, M.A, Feller, G, Gerday, C, Van Beeumen, J. | | Deposit date: | 2002-07-02 | | Release date: | 2003-03-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The Structure of a Cold-Adapted Family 8 Xylanase at 1.3 A Resolution: Structural Adaptations to Cold and Investigation of the Active Site
J.Biol.Chem., 278, 2003
|
|
1GYD
 
 | | Structure of Cellvibrio cellulosa alpha-L-arabinanase | | Descriptor: | ARABINAN ENDO-1,5-ALPHA-L-ARABINOSIDASE A | | Authors: | Nurizzo, D, Turkenburg, J.P, Charnock, S.J, Roberts, S.M, Dodson, E.J, McKie, V.A, Taylor, E.J, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2002-04-23 | | Release date: | 2002-08-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Cellvibrio japonicus alpha-L-arabinanase 43A has a novel five-blade beta-propeller fold.
Nat. Struct. Biol., 9, 2002
|
|
1GWI
 
 | | The 1.92 A structure of Streptomyces coelicolor A3(2) CYP154C1: A new monooxygenase that functionalizes macrolide ring systems | | Descriptor: | CYTOCHROME P450 154C1, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Podust, L.M, Kim, Y, Arase, M, Neely, B.A, Beck, B.J, Bach, H, Sherman, D.H, Lamb, D.C, Kelly, S.L, Waterman, M.R. | | Deposit date: | 2002-03-15 | | Release date: | 2003-01-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The 1.92 A Structure of Streptomyces Coelicolor A3(2) Cyp154C1: A New Monooxygenase that Functionalizes Macrolide Ring Systems
J.Biol.Chem., 278, 2003
|
|
2KDP
 
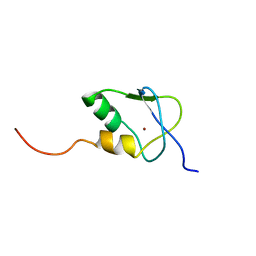 | | Solution Structure of the SAP30 zinc finger motif | | Descriptor: | Histone deacetylase complex subunit SAP30, ZINC ION | | Authors: | He, Y, Imhoff, R, Sahu, A, Radhakrishnan, I. | | Deposit date: | 2009-01-14 | | Release date: | 2009-03-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a novel zinc finger motif in the SAP30 polypeptide of the Sin3 corepressor complex and its potential role in nucleic acid recognition
Nucleic Acids Res., 37, 2009
|
|
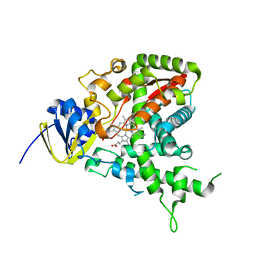
1H5Z
 
 | |
1H5Q
 
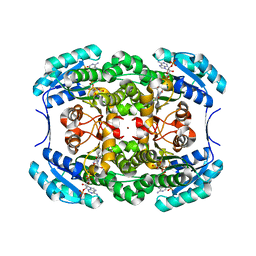 | | Mannitol dehydrogenase from Agaricus bisporus | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-DEPENDENT MANNITOL DEHYDROGENASE, NICKEL (II) ION | | Authors: | Horer, S, Stoop, J, Mooibroek, H, Baumann, U, Sassoon, J. | | Deposit date: | 2001-05-24 | | Release date: | 2001-06-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Crystallographic Structure of the Mannitol 2-Dehydrogenase Nadp+ Binary Complex from Agaricus Bisporus
J.Biol.Chem., 276, 2001
|
|
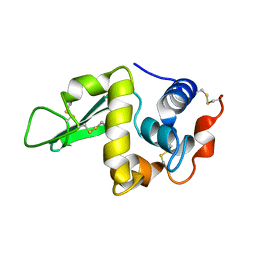
1HEN
 
 | |
1H3G
 
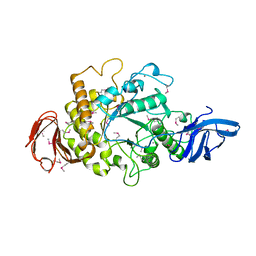 | | Cyclomaltodextrinase from Flavobacterium sp. No. 92: from DNA sequence to protein structure | | Descriptor: | CALCIUM ION, Cyclomaltodextrinase | | Authors: | Fritzsche, H.B, Schwede, T, Jelakovic, S, Schulz, G.E. | | Deposit date: | 2002-09-03 | | Release date: | 2003-08-14 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Covalent and Three-Dimensional Structure of the Cyclodextrinase from Flavobacterium Sp. No. 92.
Eur.J.Biochem., 270, 2003
|
|
2KUP
 
 | | Solution structure of the complex of the PTB domain of SNT-2 and 19-residue peptide (aa 1571-1589) of HALK | | Descriptor: | 19-residue peptide from ALK tyrosine kinase receptor, Fibroblast growth factor receptor substrate 3 | | Authors: | Li, H, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-02-24 | | Release date: | 2010-05-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the recognition of nucleophosmin-anaplastic lymphoma kinase oncoprotein by the phosphotyrosine binding domain of Suc1-associated neurotrophic factor-induced tyrosine-phosphorylated target-2
J.Struct.Funct.Genom., 11, 2010
|
|
2KHZ
 
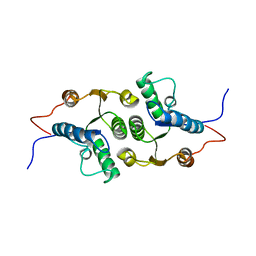 | | Solution Structure of RCL | | Descriptor: | c-Myc-responsive protein Rcl | | Authors: | Doddapaneni, K, Mahler, B, Yuan, C, Wu, Z. | | Deposit date: | 2009-04-15 | | Release date: | 2009-10-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RCL, a novel 2'-deoxyribonucleoside 5'-monophosphate N-glycosidase
J.Mol.Biol., 394, 2009
|
|
2K74
 
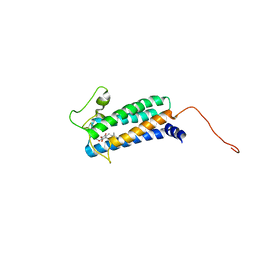 | | Solution NMR structure of DsbB-ubiquinone complex | | Descriptor: | Disulfide bond formation protein B, UBIQUINONE-2 | | Authors: | Zhou, Y, Cierpicki, T, Flores Jimenez, R.H, Lukasik, S.M, Ellena, J.F, Cafiso, D.S, Kadokura, H, Beckwith, J, Bushweller, J.H. | | Deposit date: | 2008-08-01 | | Release date: | 2008-10-07 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the integral membrane enzyme DsbB: functional insights into DsbB-catalyzed disulfide bond formation.
Mol.Cell, 31, 2008
|
|
2KJZ
 
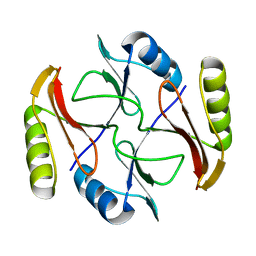 | | Solution NMR structure of protein ATC0852 from Agrobacterium tumefaciens. Northeast Structural Genomics Consortium (NESG) target AtT2. | | Descriptor: | ATC0852 | | Authors: | Lemak, A, Yee, A, Gutmanas, A, Fares, C, Garcia, M, Montelione, G, Arrowsmith, C, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-12 | | Release date: | 2009-07-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of protein ATC0852 from Agrobacterium tumefaciens
To be Published
|
|
