6RZ4
 
 | | Crystal structure of cysteinyl leukotriene receptor 1 in complex with pranlukast | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Cysteinyl leukotriene receptor 1,Soluble cytochrome b562,Cysteinyl leukotriene receptor 1, OLEIC ACID, ... | | Authors: | Luginina, A, Gusach, A, Marin, E, Mishin, A, Brouillette, R, Popov, P, Shiryaeva, A, Besserer-Offroy, E, Longpre, J.M, Lyapina, E, Ishchenko, A, Patel, N, Polovinkin, V, Safronova, N, Bogorodskiy, A, Edelweiss, E, Liu, W, Batyuk, A, Gordeliy, V, Han, G.W, Sarret, P, Katritch, V, Borshchevskiy, V, Cherezov, V. | | Deposit date: | 2019-06-12 | | Release date: | 2019-10-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-based mechanism of cysteinyl leukotriene receptor inhibition by antiasthmatic drugs.
Sci Adv, 5, 2019
|
|
7APU
 
 | | Structure of Adenylate kinase from Escherichia coli in complex with two ADP molecules refined at 1.36 A resolution. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Adenylate kinase, SODIUM ION | | Authors: | Grundstom, C, Wolf-Watz, M, Nam, K, Sauer, U.H. | | Deposit date: | 2020-10-19 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Dynamic Connection between Enzymatic Catalysis and Collective Protein Motions.
Biochemistry, 60, 2021
|
|
4ZG7
 
 | | Structural basis for inhibition of human autotaxin by four novel compounds | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl tetradecanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-({6-chloro-7-fluoro-2-methyl-1-[2-oxo-2-(spiro[cyclopropane-1,3'-indol]-1'(2'H)-yl)ethyl]-1H-indol-3-yl}sulfanyl)-2-fluorobenzoic acid, ... | | Authors: | Stein, A.J, Bain, G, Hutchinson, J.H, Evans, J.F. | | Deposit date: | 2015-04-22 | | Release date: | 2015-10-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for Inhibition of Human Autotaxin by Four Potent Compounds with Distinct Modes of Binding.
Mol.Pharmacol., 88, 2015
|
|
6RUX
 
 | | P46, an immunodominant surface protein from Mycoplasma hyopneumoniae | | Descriptor: | 46 kDa surface antigen, SODIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Guasch, A, Gonzalez-Gonzalez, L, Fita, I. | | Deposit date: | 2019-05-29 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of P46, an immunodominant surface protein from Mycoplasma hyopneumoniae: interaction with a monoclonal antibody.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
7AW7
 
 | | CCAAT-binding complex and HapX bound to Aspergillus nidulans cccA DNA | | Descriptor: | BZIP domain-containing protein, CBFD_NFYB_HMF domain-containing protein, CHLORIDE ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2020-11-06 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural insights into cooperative DNA recognition by the CCAAT-binding complex and its bZIP transcription factor HapX.
Structure, 30, 2022
|
|
8DDS
 
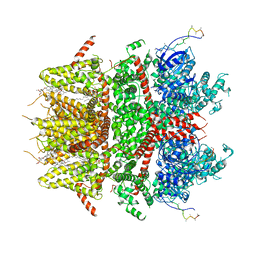 | | cryo-EM structure of TRPM3 ion channel in the presence of PIP2, state1 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, SODIUM ION, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2022-06-18 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural and functional analyses of a GPCR-inhibited ion channel TRPM3.
Neuron, 111, 2023
|
|
8DDT
 
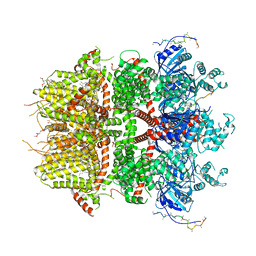 | | cryo-EM structure of TRPM3 ion channel in the presence of PIP2, state2 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, SODIUM ION, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2022-06-18 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and functional analyses of a GPCR-inhibited ion channel TRPM3.
Neuron, 111, 2023
|
|
6S08
 
 | | Crystal Structure of Properdin (TSR domains N1 & 456) | | Descriptor: | Properdin, SODIUM ION, TRIETHYLENE GLYCOL, ... | | Authors: | van den Bos, R.M, Pearce, N.M, Gros, P. | | Deposit date: | 2019-06-14 | | Release date: | 2019-09-04 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Insights Into Enhanced Complement Activation by Structures of Properdin and Its Complex With the C-Terminal Domain of C3b.
Front Immunol, 10, 2019
|
|
8D89
 
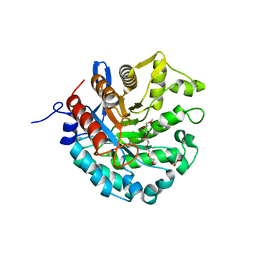 | | Crystal structure of a novel GH5 enzyme retrieved from capybara gut metagenome | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Martins, M.P, Morais, M.A.B, Murakami, M.T. | | Deposit date: | 2022-06-08 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Glycoside hydrolase subfamily GH5_57 features a highly redesigned catalytic interface to process complex hetero-beta-mannans.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7DJJ
 
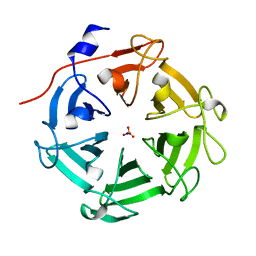 | | Structure of four truncated and mutated forms of quenching protein lumenal domains | | Descriptor: | Protein SUPPRESSOR OF QUENCHING 1, chloroplastic, SODIUM ION, ... | | Authors: | Yu, G.M, Pan, X.W, Li, M. | | Deposit date: | 2020-11-20 | | Release date: | 2022-06-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.69806433 Å) | | Cite: | Structure of Arabidopsis SOQ1 lumenal region unveils C-terminal domain essential for negative regulation of photoprotective qH.
Nat.Plants, 8, 2022
|
|
8DDU
 
 | | cryo-EM structure of TRPM3 ion channel in the presence of PIP2, state3 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, SODIUM ION, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2022-06-19 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and functional analyses of a GPCR-inhibited ion channel TRPM3.
Neuron, 111, 2023
|
|
8DDQ
 
 | | cryo-EM structure of TRPM3 ion channel in the presence of soluble Gbg, focused on channel | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, SODIUM ION, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2022-06-18 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural and functional analyses of a GPCR-inhibited ion channel TRPM3.
Neuron, 111, 2023
|
|
2F81
 
 | | HIV-1 Protease mutant L90M complexed with inhibitor TMC114 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kovalevsky, A.Y, Weber, I.T. | | Deposit date: | 2005-12-01 | | Release date: | 2006-03-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Effectiveness of Nonpeptide Clinical Inhibitor TMC-114 on HIV-1 Protease with Highly Drug Resistant Mutations D30N, I50V, and L90M.
J.Med.Chem., 49, 2006
|
|
7DJM
 
 | | Structure of four truncated and mutated forms of quenching protein | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ACETATE ION, Protein SUPPRESSOR OF QUENCHING 1, ... | | Authors: | Yu, G.M, Pan, X.W, Li, M. | | Deposit date: | 2020-11-20 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.70000112 Å) | | Cite: | Structure of Arabidopsis SOQ1 lumenal region unveils C-terminal domain essential for negative regulation of photoprotective qH.
Nat.Plants, 8, 2022
|
|
7DJK
 
 | | Structure of four truncated and mutated forms of quenching protein | | Descriptor: | CHLORIDE ION, Protein SUPPRESSOR OF QUENCHING 1, chloroplastic, ... | | Authors: | Yu, G.M, Pan, X.W, Li, M. | | Deposit date: | 2020-11-20 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.80145121 Å) | | Cite: | Structure of Arabidopsis SOQ1 lumenal region unveils C-terminal domain essential for negative regulation of photoprotective qH.
Nat.Plants, 8, 2022
|
|
8DDR
 
 | | cryo-EM structure of TRPM3 ion channel in the absence of PIP2 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, SODIUM ION, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2022-06-18 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and functional analyses of a GPCR-inhibited ion channel TRPM3.
Neuron, 111, 2023
|
|
7DJL
 
 | | Structure of four truncated and mutated forms of quenching protein | | Descriptor: | CHLORIDE ION, Protein SUPPRESSOR OF QUENCHING 1, chloroplastic, ... | | Authors: | Yu, G.M, Pan, X.W, Li, M. | | Deposit date: | 2020-11-20 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.96077824 Å) | | Cite: | Structure of Arabidopsis SOQ1 lumenal region unveils C-terminal domain essential for negative regulation of photoprotective qH.
Nat.Plants, 8, 2022
|
|
5TY0
 
 | | 2.22 Angstrom Crystal Structure of N-terminal Fragment (residues 1-419) of Elongation Factor G from Legionella pneumophila. | | Descriptor: | Elongation factor G, SODIUM ION, beta-D-glucopyranose | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Cardona-Correa, A, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-17 | | Release date: | 2016-11-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | 2.22 Angstrom Crystal Structure of N-terminal Fragment (residues 1-419) of Elongation Factor G from Legionella pneumophila.
To Be Published
|
|
7E02
 
 | |
8DDX
 
 | | cryo-EM structure of TRPM3 ion channel in complex with Gbg in the presence of PIP2, tethered by ALFA-nanobody | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2022-06-19 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural and functional analyses of a GPCR-inhibited ion channel TRPM3.
Neuron, 111, 2023
|
|
8CZA
 
 | | Crystal structure of the first bromodomain (BD1) of human BRDT bound to GXH-IV-075 | | Descriptor: | 4-[(4-{4-chloro-3-[(2-methylpropane-2-sulfonyl)amino]anilino}-5-methylpyrimidin-2-yl)amino]-2-fluoro-N-[1-(14-{3-[(2-{3-fluoro-4-[(piperidin-4-yl)carbamoyl]anilino}-5-methylpyrimidin-4-yl)amino]-5-[(2-methylpropane-2-sulfonyl)amino]phenyl}-14-oxo-4,7,10-trioxa-13-azatetradecanan-1-oyl)piperidin-4-yl]benzamide, Bromodomain testis-specific protein, SODIUM ION | | Authors: | Chan, A, Schonbrunn, E. | | Deposit date: | 2022-05-24 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Bivalent BET Bromodomain Inhibitors Confer Increased Potency and Selectivity for BRDT via Protein Conformational Plasticity.
J.Med.Chem., 65, 2022
|
|
7DTF
 
 | |
8EHE
 
 | |
4YS6
 
 | | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM CLOSTRIDIUM PHYTOFERMENTANS (Cphy_1585, TARGET EFI-511156) WITH BOUND BETA-D-GLUCOSE | | Descriptor: | CHLORIDE ION, Putative solute-binding component of ABC transporter, SODIUM ION, ... | | Authors: | Vetting, M.W, Patskovsky, Y, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-03-16 | | Release date: | 2015-04-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM CLOSTRIDIUM PHYTOFERMENTANS (Cphy_1585, TARGET EFI-511156) WITH BOUND BETA-D-GLUCOSE
To be published
|
|
2F53
 
 | | Directed Evolution of Human T-cell Receptor CDR2 residues by phage display dramatically enhances affinity for cognate peptide-MHC without apparent cross-reactivity | | Descriptor: | Beta-2-microglobulin, Cancer/testis antigen 1B, GLYCEROL, ... | | Authors: | Rizkallah, P.J, Jakobsen, B.K, Dunn, S.M, Sami, M. | | Deposit date: | 2005-11-25 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Directed evolution of human T cell receptor CDR2 residues by phage display dramatically enhances affinity for cognate peptide-MHC without increasing apparent cross-reactivity.
Protein Sci., 15, 2006
|
|
