7DEP
 
 | | S. aureus SsbB with 5-FU | | Descriptor: | 5-FLUOROURACIL, Single-stranded DNA-binding protein | | Authors: | Lin, E.S, Huang, Y.H, Huang, C.Y. | | Deposit date: | 2020-11-04 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.094 Å) | | Cite: | Crystal structure of the single-stranded DNA-binding protein SsbB in complex with the anticancer drug 5-fluorouracil: Extension of the 5-fluorouracil interactome to include the oligonucleotide/oligosaccharide-binding fold protein
Biochem.Biophys.Res.Commun., 534, 2020
|
|
1L0R
 
 | |
7D8J
 
 | | S. aureus SsbB with 5-FU | | Descriptor: | 5-FLUOROURACIL, Single-stranded DNA-binding protein | | Authors: | Lin, E.S, Huang, Y.H, Huang, C.Y. | | Deposit date: | 2020-10-08 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Crystal structure of the single-stranded DNA-binding protein SsbB in complex with the anticancer drug 5-fluorouracil: Extension of the 5-fluorouracil interactome to include the oligonucleotide/oligosaccharide-binding fold protein
Biochem.Biophys.Res.Commun., 534, 2020
|
|
8WUV
 
 | | SpCas9-MMLV RT-pegRNA-target DNA complex (elongation 16-nt) | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (5'-D(*TP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*TP*AP*CP*TP*AP*G)-3'), DNA (50-MER), ... | | Authors: | Shuto, Y, Nakagawa, R, Hoki, M, Omura, S.N, Hirano, H, Itoh, Y, Nureki, O. | | Deposit date: | 2023-10-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for pegRNA-guided reverse transcription by a prime editor.
Nature, 631, 2024
|
|
8WUU
 
 | | SpCas9-pegRNA-target DNA complex (pre-initiation) | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (34-MER), DNA (5'-D(*TP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*TP*AP*CP*TP*AP*G)-3'), ... | | Authors: | Shuto, Y, Nakagawa, R, Hoki, M, Omura, S.N, Hirano, H, Itoh, Y, Nureki, O. | | Deposit date: | 2023-10-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for pegRNA-guided reverse transcription by a prime editor.
Nature, 631, 2024
|
|
8WUS
 
 | | SpCas9-MMLV RT-pegRNA-target DNA complex (termination) | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (40-MER), DNA (5'-D(*TP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*TP*AP*CP*TP*AP*G)-3'), ... | | Authors: | Shuto, Y, Nakagawa, R, Hoki, M, Omura, S.N, Hirano, H, Itoh, Y, Nureki, O. | | Deposit date: | 2023-10-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for pegRNA-guided reverse transcription by a prime editor.
Nature, 631, 2024
|
|
8X5V
 
 | | BlCas9-sgRNA-target DNA complex | | Descriptor: | 1,2-ETHANEDIOL, BlCas9, CHLORIDE ION, ... | | Authors: | Nakane, T, Nakagawa, R, Yamashita, K, Nishimasu, H, Nureki, O. | | Deposit date: | 2023-11-19 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and engineering of Brevibacillus laterosporus Cas9.
Commun Biol, 7, 2024
|
|
2WBT
 
 | | The Structure of a Double C2H2 Zinc Finger Protein from a Hyperthermophilic Archaeal Virus in the Absence of DNA | | Descriptor: | B-129, ZINC ION | | Authors: | Eilers, B.J, Menon, S, Windham, A.B, Kraft, P, Dlakic, M, Young, M.J, Lawrence, C.M. | | Deposit date: | 2009-03-03 | | Release date: | 2010-03-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Structure of a Double C2H2 Zinc Finger Protein from a Hyperthermophilic Archaeal Virus in the Absence of DNA
To be Published
|
|
6A45
 
 | | Structure of mouse TREX2 | | Descriptor: | PHOSPHATE ION, Three prime repair exonuclease 2 | | Authors: | Hsiao, Y.Y. | | Deposit date: | 2018-06-19 | | Release date: | 2018-11-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | Structural insights into the duplex DNA processing of TREX2
Nucleic Acids Res., 46, 2018
|
|
8FZT
 
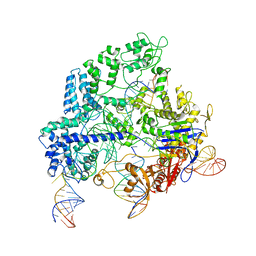 | | SpCas9 with dual-guide RNA and target DNA | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, CrE2, E2-NTS, ... | | Authors: | Korolev, S, Gagnon, K. | | Deposit date: | 2023-01-30 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | SpCas9 with dual-guide RNA and target DNA
To Be Published
|
|
2GAI
 
 | |
7DQQ
 
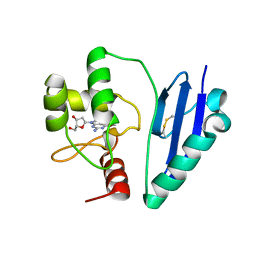 | | Crystal structure of Sulfurisphaera tokodaii O6-methylguanine methyltransferase Y91F/C120S variant in complex with O6-methyldeoxyguanosine | | Descriptor: | (2~{R},3~{S},5~{R})-5-(2-azanyl-6-methoxy-purin-9-yl)-2-(hydroxymethyl)oxolan-3-ol, Methylated-DNA--protein-cysteine methyltransferase | | Authors: | Kikuchi, M, Yamauchi, T, Iizuka, Y, Tsunoda, M. | | Deposit date: | 2020-12-24 | | Release date: | 2021-12-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Roles of the hydroxy group of tyrosine in crystal structures of Sulfurisphaera tokodaii O6-methylguanine-DNA methyltransferase.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
8A5D
 
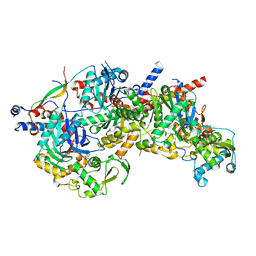 | | Structure of Arp4-Ies4-N-actin-Arp8-Ino80HSA subcomplex (A-module) of Chaetomium thermophilum INO80 | | Descriptor: | Actin, Actin related protein 4 (Arp4), Actin-related protein 8, ... | | Authors: | Kunert, F, Metzner, F.J, Eustermann, S, Jung, J, Woike, S, Schall, K, Kostrewa, D, Hopfner, K.P. | | Deposit date: | 2022-06-14 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural mechanism of extranucleosomal DNA readout by the INO80 complex.
Sci Adv, 8, 2022
|
|
1S7Z
 
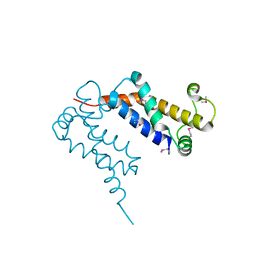 | | Structure of Ocr from Bacteriophage T7 | | Descriptor: | CESIUM ION, Gene 0.3 protein | | Authors: | Walkinshaw, M.D, Taylor, P, Sturrock, S.S, Atanasiu, C, Berg, T, Henderson, R.M, Edwardson, J.M, Dryden, D.T. | | Deposit date: | 2004-01-30 | | Release date: | 2004-02-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure of Ocr from Bacteriophage T7, a Protein that Mimics B-Form DNA
Mol.Cell, 9, 2002
|
|
8GJ9
 
 | |
6W44
 
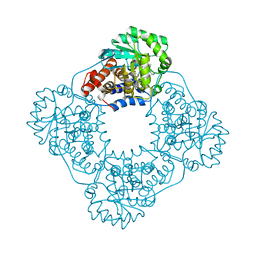 | | Crystal structure of HAO1 in complex with indazole acid inhibitor - compound 4 | | Descriptor: | 5-[methyl-[(2-propoxypyridin-3-yl)methyl]amino]-2~{H}-indazole-3-carboxylic acid, FLAVIN MONONUCLEOTIDE, Hydroxyacid oxidase 1 | | Authors: | Ferguson, A.D. | | Deposit date: | 2020-03-10 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Discovery of Novel, Potent Inhibitors of Hydroxy Acid Oxidase 1 (HAO1) Using DNA-Encoded Chemical Library Screening.
J.Med.Chem., 64, 2021
|
|
6W45
 
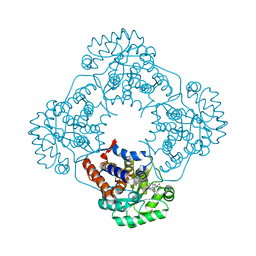 | | Crystal structure of HAO1 in complex with biaryl acid inhibitor - compound 3 | | Descriptor: | 2-chloranyl-4-[2-[[(6-chloranyl-1~{H}-indol-2-yl)carbonyl-methyl-amino]methyl]-5-fluoranyl-phenyl]benzoic acid, FLAVIN MONONUCLEOTIDE, Hydroxyacid oxidase 1 | | Authors: | Ferguson, A.D. | | Deposit date: | 2020-03-10 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of Novel, Potent Inhibitors of Hydroxy Acid Oxidase 1 (HAO1) Using DNA-Encoded Chemical Library Screening.
J.Med.Chem., 64, 2021
|
|
6W4C
 
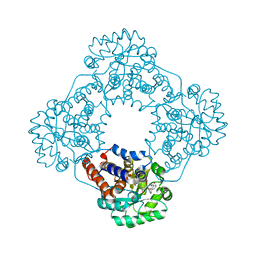 | | Crystal structure of HAO1 in complex with indazole acid inhibitor - compound 5 | | Descriptor: | 5-[[3-[3-(dimethylamino)-1,2,4-oxadiazol-5-yl]-2-oxidanyl-phenyl]methylamino]-2~{H}-indazole-3-carboxylic acid, FLAVIN MONONUCLEOTIDE, Hydroxyacid oxidase 1 | | Authors: | Ferguson, A.D. | | Deposit date: | 2020-03-10 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of Novel, Potent Inhibitors of Hydroxy Acid Oxidase 1 (HAO1) Using DNA-Encoded Chemical Library Screening.
J.Med.Chem., 64, 2021
|
|
5XH6
 
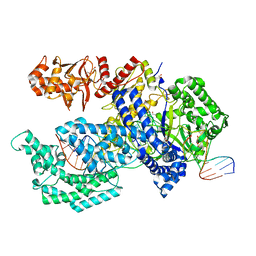 | | Crystal structure of the Acidaminococcus sp. BV3L6 Cpf1 RVR variant in complex with crRNA and target DNA (TATA PAM) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CRISPR-associated endonuclease Cpf1, ... | | Authors: | Nishimasu, H, Yamano, T, Ishitani, R, Nureki, O. | | Deposit date: | 2017-04-19 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Altered PAM Recognition by Engineered CRISPR-Cpf1
Mol. Cell, 67, 2017
|
|
1UTX
 
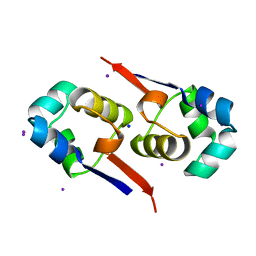 | | Regulation of Cytolysin Expression by Enterococcus faecalis: Role of CylR2 | | Descriptor: | CYLR2, IODIDE ION, SODIUM ION | | Authors: | Razeto, A, Rumpel, S, Pillar, C.M, Gilmore, M.S, Becker, S, Zweckstetter, M. | | Deposit date: | 2003-12-12 | | Release date: | 2004-09-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and DNA-Binding Properties of the Cytolysin Regulator CylR2 from Enterococcus Faecalis
Embo J., 23, 2004
|
|
6VYC
 
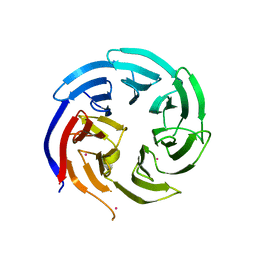 | | Crystal structure of WD-repeat domain of human WDR91 | | Descriptor: | UNKNOWN ATOM OR ION, WD repeat-containing protein 91 | | Authors: | Halabelian, L, Hutchinson, A, Li, Y, Seitova, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-26 | | Release date: | 2020-03-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of a First-in-Class Small-Molecule Ligand for WDR91 Using DNA-Encoded Chemical Library Selection Followed by Machine Learning.
J.Med.Chem., 66, 2023
|
|
2ZIE
 
 | | Crystal Structure of TTHA0409, Putatative DNA Modification Methylase from Thermus thermophilus HB8- Selenomethionine derivative | | Descriptor: | Putative modification methylase | | Authors: | Morita, R, Ishikawa, H, Nakagawa, N, Masui, R, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-02-15 | | Release date: | 2008-07-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a putative DNA methylase TTHA0409 from Thermus thermophilus HB8
Proteins, 73, 2008
|
|
7E1P
 
 | | Crystal structure of Sulfurisphaera tokodaii O6-methylguanine methyltransferase C120S variant in complex with O6-methyldeoxyguanosine | | Descriptor: | (2~{R},3~{S},5~{R})-5-(2-azanyl-6-methoxy-purin-9-yl)-2-(hydroxymethyl)oxolan-3-ol, Methylated-DNA--protein-cysteine methyltransferase, SULFATE ION | | Authors: | Kikuchi, M, Yamauchi, T, Iizuka, Y, Tsunoda, M. | | Deposit date: | 2021-02-03 | | Release date: | 2021-12-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Roles of the hydroxy group of tyrosine in crystal structures of Sulfurisphaera tokodaii O6-methylguanine-DNA methyltransferase.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
2VKG
 
 | | COMPLEXES OF DODECIN WITH FLAVIN AND FLAVIN-LIKE LIGANDS | | Descriptor: | CHLORIDE ION, DODECIN, MAGNESIUM ION, ... | | Authors: | Grininger, M, Noell, G, Trawoeger, S, Sinner, E, Oesterhelt, D. | | Deposit date: | 2007-12-19 | | Release date: | 2008-09-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Electrochemical Switching of the Flavoprotein Dodecin at Gold Surfaces Modified by Flavin-DNA Hybrid Linkers
Biointerphases, 3, 2008
|
|
5ET9
 
 | | Racemic crystal structures of Pribnow box consensus promoter sequence (P21/n) | | Descriptor: | BARIUM ION, Pribnow box consensus sequence- template strand, Pribnow box non-template strand | | Authors: | Mandal, P.K, Collie, G.W, Kauffmann, B, Srivastava, S.C, Huc, I. | | Deposit date: | 2015-11-17 | | Release date: | 2016-05-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure elucidation of the Pribnow box consensus promoter sequence by racemic DNA crystallography.
Nucleic Acids Res., 44, 2016
|
|
