1P9R
 
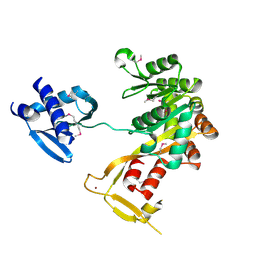 | | Crystal Structure of Vibrio cholerae putative NTPase EpsE | | Descriptor: | CHLORIDE ION, General secretion pathway protein E, ZINC ION | | Authors: | Robien, M.A, Krumm, B.E, Sandkvist, M, Hol, W.G.J. | | Deposit date: | 2003-05-12 | | Release date: | 2003-10-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the extracellular protein secretion NTPase EpsE of Vibrio cholerae
J.Mol.Biol., 333, 2003
|
|
4UPB
 
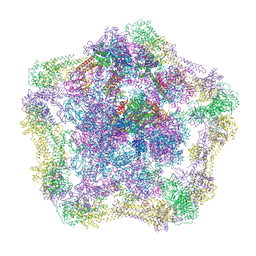 | | Electron cryo-microscopy of the complex formed between the hexameric ATPase RavA and the decameric inducible decarboxylase LdcI | | Descriptor: | ATPASE RAVA, LYSINE DECARBOXYLASE, INDUCIBLE | | Authors: | Malet, H, Liu, K, El Bakkouri, M, Chan, S.W.S, Effantin, G, Bacia, M, Houry, W.A, Gutsche, I. | | Deposit date: | 2014-06-15 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Assembly Principles of a Unique Cage Formed by Hexameric and Decameric E. Coli Proteins.
Elife, 3, 2014
|
|
3JS6
 
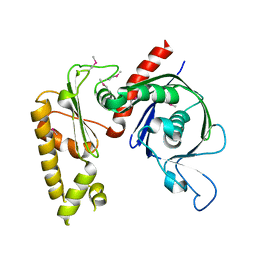 | | Crystal structure of apo psk41 parM protein | | Descriptor: | Uncharacterized ParM protein | | Authors: | Schumacher, M.A, Xu, W, Firth, N. | | Deposit date: | 2009-09-09 | | Release date: | 2010-01-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and filament dynamics of the pSK41 actin-like ParM protein: implications for plasmid DNA segregation.
J.Biol.Chem., 285, 2010
|
|
4UPF
 
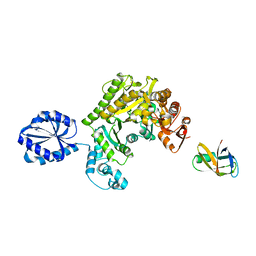 | | Assembly principles of the unique cage formed by the ATPase RavA hexamer and the lysine decarboxylase LdcI decamer | | Descriptor: | ATPASE RAVA, LYSINE DECARBOXYLASE, INDUCIBLE | | Authors: | Malet, H, Liu, K, El Bakkouri, M, Chan, S.W.S, Effantin, G, Bacia, M, Houry, W.A, Gutsche, I. | | Deposit date: | 2014-06-16 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Assembly Principles of a Unique Cage Formed by Hexameric and Decameric E. Coli Proteins.
Elife, 3, 2014
|
|
3K5X
 
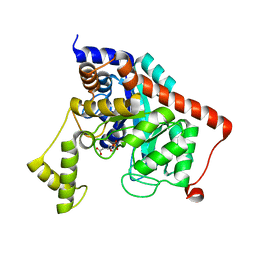 | | Crystal structure of dipeptidase from Streptomics coelicolor complexed with phosphinate pseudodipeptide L-Ala-D-Asp at 1.4A resolution. | | Descriptor: | Dipeptidase, ZINC ION, phosphinate pseudodipeptide L-Ala-D-Asp | | Authors: | Fedorov, A.A, Fedorov, E.V, Cummings, J, Raushel, F.M, Almo, S.C. | | Deposit date: | 2009-10-08 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure, mechanism, and substrate profile for Sco3058: the closest bacterial homologue to human renal dipeptidase .
Biochemistry, 49, 2010
|
|
3HQF
 
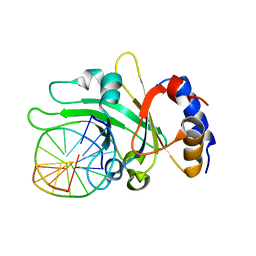 | | Crystal structure of restriction endonuclease EcoRII N-terminal effector-binding domain in complex with cognate DNA | | Descriptor: | 5'-D(*CP*GP*CP*CP*AP*GP*GP*GP*C)-3', 5'-D(*GP*CP*CP*CP*TP*GP*GP*CP*G)-3', Restriction endonuclease | | Authors: | Golovenko, D, Manakova, E, Grazulis, S, Tamulaitiene, G, Siksnys, V. | | Deposit date: | 2009-06-06 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural mechanisms for the 5'-CCWGG sequence recognition by the N- and C-terminal domains of EcoRII.
Nucleic Acids Res., 37, 2009
|
|
3HQG
 
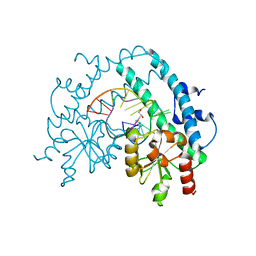 | | Crystal structure of restriction endonuclease EcoRII catalytic C-terminal domain in complex with cognate DNA | | Descriptor: | 5'-D(*TP*AP*GP*CP*CP*TP*GP*GP*TP*CP*GP*A)-3', 5'-D(*TP*CP*GP*AP*CP*CP*AP*GP*GP*CP*TP*A)-3', GLYCEROL, ... | | Authors: | Golovenko, D, Manakova, E, Grazulis, S, Tamulaitiene, G, Siksnys, V. | | Deposit date: | 2009-06-06 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural mechanisms for the 5'-CCWGG sequence recognition by the N- and C-terminal domains of EcoRII.
Nucleic Acids Res., 37, 2009
|
|
4ELD
 
 | |
1Q51
 
 | | Crystal Structure of Mycobacterium tuberculosis MenB in Complex with Acetoacetyl-Coenzyme A, a Key Enzyme in Vitamin K2 Biosynthesis | | Descriptor: | ACETOACETYL-COENZYME A, menB | | Authors: | Truglio, J.J, Theis, K, Feng, Y, Gajda, R, Machutta, C, Tonge, P.J, Kisker, C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-08-05 | | Release date: | 2004-01-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis MenB, a key enzyme in vitamin K2 biosynthesis.
J.Biol.Chem., 278, 2003
|
|
3K4Y
 
 | |
3K56
 
 | |
3K4O
 
 | |
1N4D
 
 | |
1Q52
 
 | | Crystal Structure of Mycobacterium tuberculosis MenB, a Key Enzyme in Vitamin K2 Biosynthesis | | Descriptor: | menB | | Authors: | Truglio, J.J, Theis, K, Feng, Y, Gajda, R, Machutta, C, Tonge, P.J, Kisker, C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-08-05 | | Release date: | 2004-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis MenB, a key enzyme in vitamin K2 biosynthesis.
J.Biol.Chem., 278, 2003
|
|
8J9F
 
 | | Structure of STG-hydrolyzing beta-glucosidase 1 (PSTG1) | | Descriptor: | Beta-glucosidase, GLYCEROL | | Authors: | Yanai, T, Imaizumi, R, Takahashi, Y, Katsumura, E, Yamamoto, M, Nakayama, T, Yamashita, S, Takeshita, K, Sakai, N, Matsuura, H. | | Deposit date: | 2023-05-03 | | Release date: | 2024-04-10 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural insights into a bacterial beta-glucosidase capable of degrading sesaminol triglucoside to produce sesaminol: toward the understanding of the aglycone recognition mechanism by the C-terminal lid domain.
J.Biochem., 174, 2023
|
|
5FFS
 
 | | Crystal Structure of Surfactant Protein-A Y164A Mutant | | Descriptor: | CALCIUM ION, Pulmonary surfactant-associated protein A | | Authors: | Goh, B.C, Wu, H, Rynkiewicz, M.J, Schulten, K, Seaton, B.A, McCormack, F.X. | | Deposit date: | 2015-12-18 | | Release date: | 2016-07-06 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Elucidation of Lipid Binding Sites on Lung Surfactant Protein A Using X-ray Crystallography, Mutagenesis, and Molecular Dynamics Simulations.
Biochemistry, 55, 2016
|
|
6VRS
 
 | | Single particle reconstruction of glucose isomerase from Streptomyces rubiginosus based on data acquired in the presence of substantial aberrations | | Descriptor: | MANGANESE (II) ION, xylose isomerase | | Authors: | Bromberg, R, Guo, Y, Borek, D, Otwinowski, Z. | | Deposit date: | 2020-02-09 | | Release date: | 2020-02-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | High-resolution cryo-EM reconstructions in the presence of substantial aberrations
Iucrj, 7, 2020
|
|
5MR3
 
 | | Crystal structure of red abalone egg VERL repeat 2 with linker in complex with sperm lysin at 1.8 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Egg-lysin, ... | | Authors: | Nishimura, K, Raj, I, Sadat Al-Hosseini, H, De Sanctis, D, Jovine, L. | | Deposit date: | 2016-12-21 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Egg Coat-Sperm Recognition at Fertilization.
Cell, 169, 2017
|
|
6VSC
 
 | |
5FJI
 
 | | Three-dimensional structures of two heavily N-glycosylated Aspergillus sp. Family GH3 beta-D-glucosidases | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-GLUCOSIDASE, ... | | Authors: | Agirre, J, Ariza, A, Offen, W.A, Turkenburg, J.P, Roberts, S.M, McNicholas, S, Harris, P.V, McBrayer, B, Dohnalek, J, Cowtan, K.D, Davies, G.J, Wilson, K.S. | | Deposit date: | 2015-10-09 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Three-Dimensional Structures of Two Heavily N-Glycosylated Aspergillus Sp. Family Gh3 Beta-D-Glucosidases
Acta Crystallogr.,Sect.D, 72, 2016
|
|
6VI8
 
 | | Observing a ring-cleaving dioxygenase in action through a crystalline lens - a superoxo bound structure | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-HYDROXYANTHRANILIC ACID, 3-hydroxyanthranilate 3,4-dioxygenase, ... | | Authors: | Wang, Y, Liu, F, Yang, Y, Liu, A. | | Deposit date: | 2020-01-12 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Observing 3-hydroxyanthranilate-3,4-dioxygenase in action through a crystalline lens.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5FES
 
 | | HydE from T. maritima in complex with (2R,4R)-MeSeTDA | | Descriptor: | (2~{R},4~{R})-2-methyl-1,3-selenazolidine-2,4-dicarboxylic acid, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CHLORIDE ION, ... | | Authors: | Rohac, R, Amara, P, Benjdia, A, Martin, L, Ruffie, P, Favier, A, Berteau, O, Mouesca, J.M, Fontecilla-Camps, J.C, Nicolet, Y. | | Deposit date: | 2015-12-17 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Carbon-sulfur bond-forming reaction catalysed by the radical SAM enzyme HydE.
Nat.Chem., 8, 2016
|
|
5FF2
 
 | | HydE from T. maritima in complex with (2R,4R)-TDA | | Descriptor: | (2~{R},4~{R})-1,3-thiazolidine-2,4-dicarboxylic acid, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CHLORIDE ION, ... | | Authors: | Rohac, R, Amara, P, Benjdia, A, Martin, L, Ruffie, P, Favier, A, Berteau, O, Mouesca, J.M, Fontecilla-Camps, J.C, Nicolet, Y. | | Deposit date: | 2015-12-17 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Carbon-sulfur bond-forming reaction catalysed by the radical SAM enzyme HydE.
Nat.Chem., 8, 2016
|
|
6VIA
 
 | | Observing a ring-cleaving dioxygenase in action through a crystalline lens - a seven-membered lactone bound structure | | Descriptor: | (2R,3E)-2-hydroxy-3-imino-2,3-dihydrooxepine-4-carboxylic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-hydroxyanthranilate 3,4-dioxygenase, ... | | Authors: | Wang, Y, Liu, F, Yang, Y, Liu, A. | | Deposit date: | 2020-01-12 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.591 Å) | | Cite: | Observing 3-hydroxyanthranilate-3,4-dioxygenase in action through a crystalline lens.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2CV4
 
 | | Crystal Structure of an Archaeal Peroxiredoxin from the Aerobic Hyperthermophilic Crenarchaeon Aeropyrum pernix K1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ISOPROPYL ALCOHOL, peroxiredoxin | | Authors: | Mizohata, E, Sakai, H, Fusatomi, E, Terada, T, Murayama, K, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-31 | | Release date: | 2005-06-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of an Archaeal Peroxiredoxin from the Aerobic Hyperthermophilic Crenarchaeon Aeropyrum pernix K1
J.Mol.Biol., 354, 2005
|
|
