8ORP
 
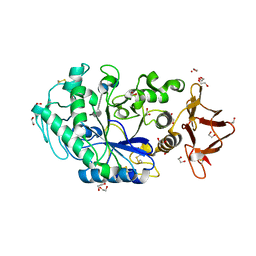 | | Crystal structure of Drosophila melanogaster alpha-amylase in complex with the inhibitor acarbose | | Descriptor: | 1,2-ETHANEDIOL, 4,6-dideoxy-4-{[(1S,2S,3S,4R,5R)-2,3,4-trihydroxy-5-(hydroxymethyl)cyclohexyl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 4,6-dideoxy-4-{[(1S,2S,3S,4R,5R)-2,3,4-trihydroxy-5-(hydroxymethyl)cyclohexyl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Aghajari, N, Haser, R. | | Deposit date: | 2023-04-16 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Functional Characterization of Drosophila melanogaster alpha-Amylase.
Molecules, 28, 2023
|
|
6OE6
 
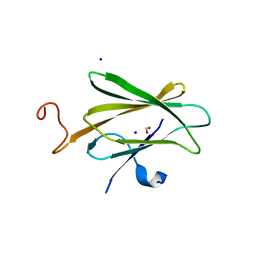 | |
5BY3
 
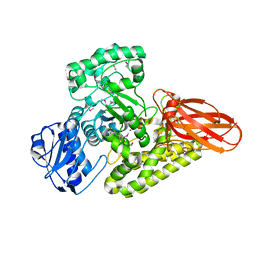 | | A novel family GH115 4-O-Methyl-alpha-glucuronidase, BtGH115A, with specificity for decorated arabinogalactans | | Descriptor: | BtGH115A, NICKEL (II) ION, SULFATE ION | | Authors: | Lammerts van Bueren, A, Davies, G.J, Turkenburg, J.P. | | Deposit date: | 2015-06-10 | | Release date: | 2015-07-22 | | Last modified: | 2015-12-09 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structural and Functional Characterization of a Novel Family GH115 4-O-Methyl-alpha-Glucuronidase with Specificity for Decorated Arabinogalactans.
J.Mol.Biol., 427, 2015
|
|
4FQE
 
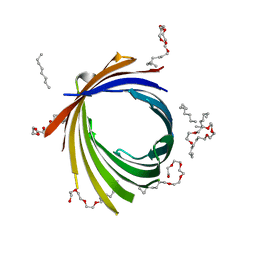 | | KdgM porin | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, N-OCTANE, Oligogalacturonate-specific porin kdgM | | Authors: | Hutter, C, Peneff, C.M, Wirth, C, Schirmer, T. | | Deposit date: | 2012-06-25 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure of the oligogalacturonate-specific KdgM porin.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6ER1
 
 | | Crystal structure of BTB-domain of CP190 from D.melanogaster at high resolution | | Descriptor: | Centrosome-associated zinc finger protein CP190, PHOSPHATE ION | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Bonchuk, A.N, Kachalova, G.S, Georgiev, P.G, Popov, V.O. | | Deposit date: | 2017-10-16 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Purification, Isolation, Crystallization, and Preliminary X-ray Diffraction Study of the BTB Domain of the Centrosomal Protein 190 from Drosophila Melanogaster
Crystallography Reports, 62, 2017
|
|
8OR6
 
 | |
1CO6
 
 | |
7PX0
 
 | |
7PV5
 
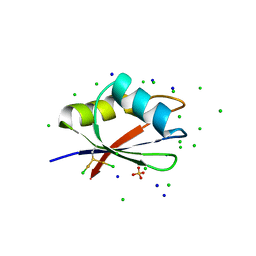 | | RBD domain of D. melanogaster tRNA (uracil-5-)-methyltransferase homolog A (TRMT2A) | | Descriptor: | CHLORIDE ION, FI05218p, SODIUM ION, ... | | Authors: | Witzenberger, M, Janowski, R, Niessing, D. | | Deposit date: | 2021-10-01 | | Release date: | 2023-04-19 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the RNA-recognition motif of Drosophila melanogaster tRNA (uracil-5-)-methyltransferase homolog A
Acta Crystallogr.,Sect.F, 80, 2024
|
|
6H1A
 
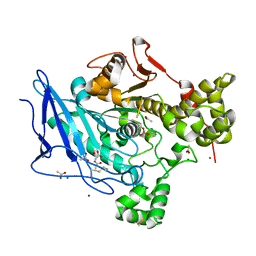 | |
5HC9
 
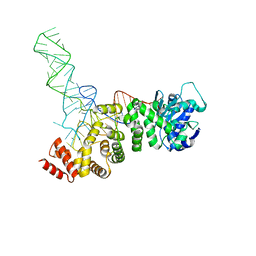 | |
2LVB
 
 | | Solution NMR Structure DE NOVO DESIGNED PFK fold PROTEIN, Northeast Structural Genomics Consortium (NESG) Target OR250 | | Descriptor: | DE NOVO DESIGNED PFK fold PROTEIN | | Authors: | Liu, G, Koga, N, Koga, R, Xiao, R, Hamilton, K, Kohan, E, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-06-30 | | Release date: | 2012-08-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Principles for designing ideal protein structures.
Nature, 491, 2012
|
|
7S37
 
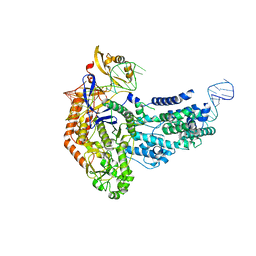 | | Cas9:sgRNA (S. pyogenes) in the open-protein conformation | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, Single-guide RNA | | Authors: | Cofsky, J.C, Soczek, K.M, Knott, G.J, Nogales, E, Doudna, J.A. | | Deposit date: | 2021-09-04 | | Release date: | 2022-04-20 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | CRISPR-Cas9 bends and twists DNA to read its sequence.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7S3H
 
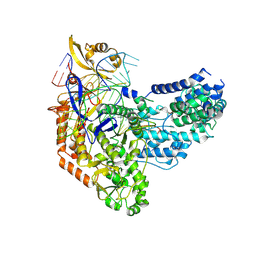 | | Cas9:sgRNA:DNA (S. pyogenes) with 0 RNA:DNA base pairs, open-protein/linear-DNA conformation | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, Non-target DNA strand, Single-guide RNA, ... | | Authors: | Cofsky, J.C, Soczek, K.M, Knott, G.J, Nogales, E, Doudna, J.A. | | Deposit date: | 2021-09-06 | | Release date: | 2022-04-20 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | CRISPR-Cas9 bends and twists DNA to read its sequence.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7S36
 
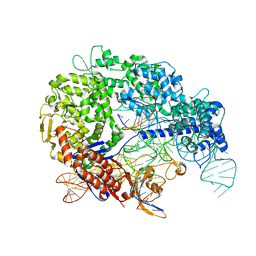 | | Cas9:sgRNA:DNA (S. pyogenes) with 0 RNA:DNA base pairs, closed-protein/bent-DNA conformation | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, Non-target DNA strand, Single-guide RNA, ... | | Authors: | Cofsky, J.C, Soczek, K.M, Knott, G.J, Nogales, E, Doudna, J.A. | | Deposit date: | 2021-09-04 | | Release date: | 2022-04-20 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | CRISPR-Cas9 bends and twists DNA to read its sequence.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7S38
 
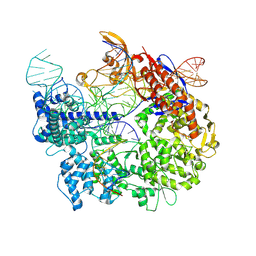 | | Cas9:sgRNA:DNA (S. pyogenes) forming a 3-base-pair R-loop | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, Non-target DNA strand, Single-guide RNA, ... | | Authors: | Cofsky, J.C, Soczek, K.M, Knott, G.J, Nogales, E, Doudna, J.A. | | Deposit date: | 2021-09-04 | | Release date: | 2022-04-20 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | CRISPR-Cas9 bends and twists DNA to read its sequence.
Nat.Struct.Mol.Biol., 29, 2022
|
|
2LV8
 
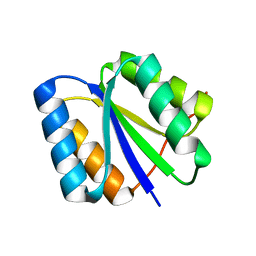 | | Solution NMR Structure de novo designed rossmann 2x2 fold protein, Northeast Structural Genomics Consortium (NESG) Target OR16 | | Descriptor: | De novo designed rossmann 2x2 fold protein | | Authors: | Liu, G, Koga, R, Koga, N, Xiao, R, Pederson, K, Hamilton, K, Ciccosanti, C, Acton, T.B, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-06-29 | | Release date: | 2012-08-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Principles for designing ideal protein structures.
Nature, 491, 2012
|
|
6P5A
 
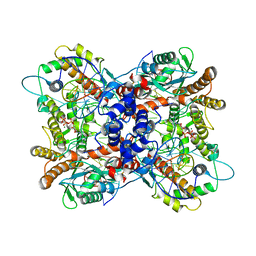 | | Drosophila P element transposase strand transfer complex | | Descriptor: | DNA (44-MER), DNA (5'-D(P*AP*GP*GP*TP*GP*GP*TP*CP*CP*CP*GP*TP*CP*GP*G)-3'), DNA (5'-D(P*CP*GP*AP*AP*CP*TP*AP*TP*A)-3'), ... | | Authors: | Kellogg, E.H, Nogales, E, Ghanim, G, Rio, D.C. | | Deposit date: | 2019-05-30 | | Release date: | 2019-10-30 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of a P element transposase-DNA complex reveals unusual DNA structures and GTP-DNA contacts.
Nat.Struct.Mol.Biol., 26, 2019
|
|
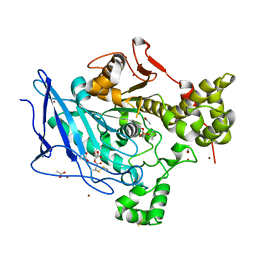
6H18
 
 | |
6H0V
 
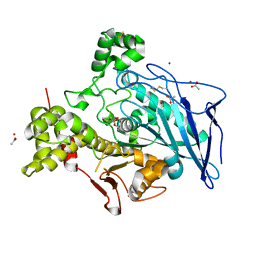 | |
6PE2
 
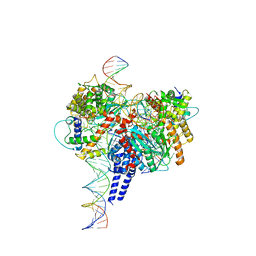 | | Drosophila P element transposase strand transfer complex | | Descriptor: | DNA (27-MER), DNA (5'-D(P*CP*GP*AP*AP*CP*TP*AP*TP*A)-3'), DNA (56-MER), ... | | Authors: | Kellogg, E.H, Nogales, E, Ghanim, G, Rio, D.C. | | Deposit date: | 2019-06-19 | | Release date: | 2019-10-30 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of a P element transposase-DNA complex reveals unusual DNA structures and GTP-DNA contacts.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6PZJ
 
 | |
4P0O
 
 | | Cleaved Serpin 42Da | | Descriptor: | Serine protease inhibitor 4, isoform B | | Authors: | Ellisdon, A.M, Whisstock, J.C. | | Deposit date: | 2014-02-21 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High resolution structure of cleaved Serpin 42 Da from Drosophila melanogaster.
Bmc Struct.Biol., 14, 2014
|
|
8JSY
 
 | |
8JSV
 
 | |
