6RR9
 
 | | DNA/RNA binding protein | | Descriptor: | GLYCEROL, SULFATE ION, Schlafen family member 5, ... | | Authors: | Huber, E, Lammens, K. | | Deposit date: | 2019-05-17 | | Release date: | 2020-07-08 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3.432 Å) | | Cite: | Structural and biochemical characterization of human Schlafen 5.
Nucleic Acids Res., 2022
|
|
5IJ2
 
 | | SrpA adhesin in complex with sialyllactosamine | | Descriptor: | ACETATE ION, CALCIUM ION, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Iverson, T.M. | | Deposit date: | 2016-03-01 | | Release date: | 2017-02-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.683 Å) | | Cite: | Structures of the Streptococcus sanguinis SrpA Binding Region with Human Sialoglycans Suggest Features of the Physiological Ligand.
Biochemistry, 2016
|
|
6RSD
 
 | |
4PKK
 
 | |
6RSU
 
 | | TBK1 in complex with Inhibitor compound 35 | | Descriptor: | 3,3,3-tris(fluoranyl)-1-[4-[(1~{R})-1-[2-[[(2~{S})-5-(5-propan-2-yloxypyrimidin-4-yl)-2,3-dihydro-1~{H}-benzimidazol-2-yl]amino]pyridin-4-yl]ethyl]piperazin-1-yl]propan-1-one, Serine/threonine-protein kinase TBK1 | | Authors: | Panne, D, Hillig, R.C, Rengachari, S. | | Deposit date: | 2019-05-22 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Discovery of BAY-985, a Highly Selective TBK1/IKK epsilon Inhibitor.
J.Med.Chem., 63, 2020
|
|
6RTT
 
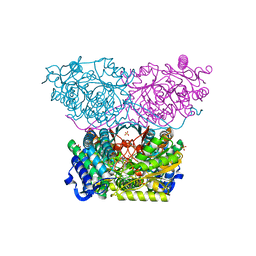 | | Piperideine-6-carboxylate dehydrogenase from Streptomyces clavuligerus complexed with picolinic acid | | Descriptor: | GLYCEROL, PYRIDINE-2-CARBOXYLIC ACID, SULFATE ION, ... | | Authors: | Hasse, D, Huelsemann, J, Carlsson, G, Andersson, I. | | Deposit date: | 2019-05-26 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and mechanism of piperideine-6-carboxylate dehydrogenase from Streptomyces clavuligerus.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6RTB
 
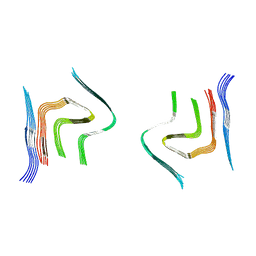 | | cryo-em structure of alpha-synuclein fibril polymorph 2B | | Descriptor: | Alpha-synuclein | | Authors: | Guerrero-Ferreira, R, Taylor, N.M.I, Arteni, A.A, Melki, R, Meier, B.H, Bockmann, A, Bousset, L, Stahlberg, H. | | Deposit date: | 2019-05-22 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Two new polymorphic structures of human full-length alpha-synuclein fibrils solved by cryo-electron microscopy.
Elife, 8, 2019
|
|
4UU3
 
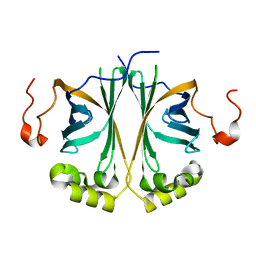 | | Ferulic acid decarboxylase from Enterobacter sp. | | Descriptor: | FERULIC ACID DECARBOXYLASE | | Authors: | Hromic, A, Pavkov-Keller, T, Steinkellner, G, Lyskowski, A, Wuensch, C, Gross, J, Fuchs, M, Fauland, K, Glueck, S.M, Faber, K, Gruber, K. | | Deposit date: | 2014-07-24 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Regioselective Enzymatic Beta-Carboxylation of Para-Hydroxy-Styrene Derivatives Catalyzed by Phenolic Acid Decarboxylases.
Adv. Synth. Catal., 357, 2015
|
|
6RWD
 
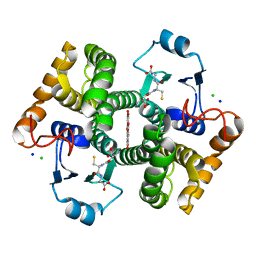 | | Crystal structure of SjGST in complex with GSH and ellagic acid at 1.53 Angstrom resolution | | Descriptor: | 2,3,7,8-tetrahydroxychromeno[5,4,3-cde]chromene-5,10-dione, CHLORIDE ION, GLUTATHIONE, ... | | Authors: | Olfsen, J, Pandian, R, Sayed, Y, Dirr, H.W, Achilonu, I.A. | | Deposit date: | 2019-06-04 | | Release date: | 2019-06-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Molecular basis of inhibition of Schistosoma japonicum glutathione transferase by ellagic acid: Insights into biophysical and structural studies.
Mol.Biochem.Parasitol., 240, 2020
|
|
6RUG
 
 | |
5I29
 
 | | TAF1(2) bound to a pyrrolopyridone compound | | Descriptor: | CALCIUM ION, N,N-dimethyl-3-(6-methyl-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridin-4-yl)benzamide, Transcription initiation factor TFIID subunit 1 | | Authors: | Tang, Y, Poy, F, Bellon, S.F. | | Deposit date: | 2016-02-08 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Diving into the Water: Inducible Binding Conformations for BRD4, TAF1(2), BRD9, and CECR2 Bromodomains.
J.Med.Chem., 59, 2016
|
|
5IM5
 
 | | Crystal structure of designed two-component self-assembling icosahedral cage I53-40 | | Descriptor: | Designed Keto-hydroxyglutarate-aldolase/keto-deoxy-phosphogluconate aldolase, Designed Riboflavin synthase | | Authors: | Liu, Y.A, Cascio, D, Sawaya, M.R, Bale, J.B, Collazo, M.J, Thomas, C, Sheffler, W, King, N.P, Baker, D, Yeates, T.O. | | Deposit date: | 2016-03-05 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.699 Å) | | Cite: | Accurate design of megadalton-scale two-component icosahedral protein complexes.
Science, 353, 2016
|
|
6RXC
 
 | | Leishmania major pteridine reductase 1 (LmPTR1) in complex with inhibitor 4 (NMT-C0026) | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pteridine reductase 1, methyl 1-[4-[[2,4-bis(azanyl)pteridin-6-yl]methyl-(3-oxidanylpropyl)amino]phenyl]carbonylpiperidine-4-carboxylate | | Authors: | Di Pisa, F, Dello Iacono, L, Pozzi, C, Mangani, S. | | Deposit date: | 2019-06-07 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Multitarget, Selective Compound Design Yields Potent Inhibitors of a Kinetoplastid Pteridine Reductase 1.
J.Med.Chem., 65, 2022
|
|
6RZ8
 
 | | Crystal structure of the human cysteinyl leukotriene receptor 2 in complex with ONO-2080365 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2~{S})-8-[[4-[4-[2,3-bis(fluoranyl)phenoxy]butoxy]-2-fluoranyl-phenyl]carbonylamino]-4-(4-oxidanyl-4-oxidanylidene-but yl)-2,3-dihydro-1,4-benzoxazine-2-carboxylic acid, Cysteinyl leukotriene receptor 2,Soluble cytochrome b562,Cysteinyl leukotriene receptor 2, ... | | Authors: | Gusach, A, Luginina, A, Marin, E, Brouillette, R.L, Besserer-Offroy, E, Longpre, J.M, Ishchenko, A, Popov, P, Fujimoto, T, Maruyama, T, Stauch, B, Ergasheva, M, Romanovskaya, D, Stepko, A, Kovalev, K, Shevtsov, M, Gordeliy, V, Han, G.W, Sarret, P, Katritch, V, Borshchevskiy, V, Mishin, A, Cherezov, V. | | Deposit date: | 2019-06-12 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of ligand selectivity and disease mutations in cysteinyl leukotriene receptors.
Nat Commun, 10, 2019
|
|
6RY2
 
 | | Crystal structure of Dfg5 from Chaetomium thermophilum in complex with alpha-1,2-mannobiose | | Descriptor: | ACETATE ION, CALCIUM ION, Mannan endo-1,6-alpha-mannosidase, ... | | Authors: | Essen, L.-O, Vogt, M.S. | | Deposit date: | 2019-06-10 | | Release date: | 2020-08-12 | | Last modified: | 2020-09-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural base for the transfer of GPI-anchored glycoproteins into fungal cell walls.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6RZE
 
 | | Crystal structure of E. coli Adenylate kinase R119A mutant | | Descriptor: | Adenylate kinase, CHLORIDE ION, SODIUM ION | | Authors: | Grundstrom, C, Rogne, P, Wolf-Watz, M, Sauer-Eriksson, A.E. | | Deposit date: | 2019-06-13 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Nucleation of an Activating Conformational Change by a Cation-pi Interaction.
Biochemistry, 58, 2019
|
|
7S51
 
 | | Structure of C208A Sortase A from Streptococcus pyogenes bound to LPATA peptide | | Descriptor: | LEU-PRO-ALA-THR-ALA, Sortase | | Authors: | Johnson, D.A, Svendsen, J.E, Antos, J.M, Amacher, J.F. | | Deposit date: | 2021-09-09 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structures of Streptococcus pyogenes class A sortase in complex with substrate and product mimics provide key details of target recognition.
J.Biol.Chem., 298, 2022
|
|
6RZI
 
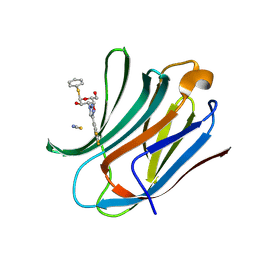 | | Galectin-3C in complex with trifluoroaryltriazole monothiogalactoside derivative:1(Hydrogen) | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{R})-2-(hydroxymethyl)-6-phenylsulfanyl-4-[4-[3,4,5-tris(fluoranyl)phenyl]-1,2,3-triazol-1-yl]oxane-3,5-diol, Galectin-3, THIOCYANATE ION | | Authors: | Kumar, R, Verteramo, M.L, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2019-06-13 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.095 Å) | | Cite: | Thermodynamic studies of halogen-bond interactions in galectin-3
to be published
|
|
5IOD
 
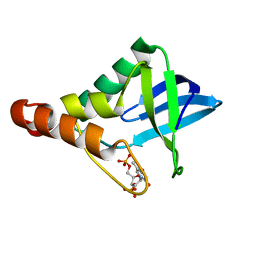 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V66H/V99E at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Robinson, A.C, Schlessman, J.L, Theodoru, A, Garcia-Moreno E, B. | | Deposit date: | 2016-03-08 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS V66H/V99E at cryogenic temperature
To be Published
|
|
7S4O
 
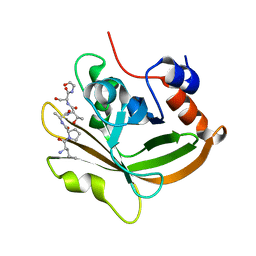 | | Structure of C208A Sortase A from Streptococcus pyogenes bound to LPATS peptide | | Descriptor: | LEU-PRO-ALA-THR-SER-GLY, Sortase | | Authors: | Johnson, D.A, Svendsen, J.E, Antos, J.M, Amacher, J.F. | | Deposit date: | 2021-09-09 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.396 Å) | | Cite: | Structures of Streptococcus pyogenes class A sortase in complex with substrate and product mimics provide key details of target recognition.
J.Biol.Chem., 298, 2022
|
|
6RZJ
 
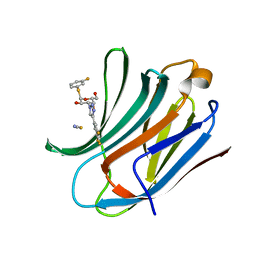 | | Galectin-3C in complex with trifluoroaryltriazole monothiogalactoside derivative:2(Fluorine) | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{R})-2-(3-fluorophenyl)sulfanyl-6-(hydroxymethyl)-4-[4-[3,4,5-tris(fluoranyl)phenyl]-1,2,3-triazol-1-yl]oxane-3,5-diol, Galectin-3, THIOCYANATE ION | | Authors: | Kumar, R, Verteramo, M.L, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2019-06-13 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.097 Å) | | Cite: | Thermodynamic studies of halogen-bond interactions in galectin-3
to be published
|
|
4PNG
 
 | | Glutathione S-transferase from Drosophila melanogaster - isozyme E7 | | Descriptor: | L-GAMMA-GLUTAMYL-3-SULFINO-L-ALANYLGLYCINE, LD04004p | | Authors: | Scian, M, Le Trong, I, Mannervik, B, Atkins, W.M, Stenkamp, R.E. | | Deposit date: | 2014-05-23 | | Release date: | 2015-05-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Comparison of epsilon- and delta-class glutathione S-transferases: the crystal structures of the glutathione S-transferases DmGSTE6 and DmGSTE7 from Drosophila melanogaster.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6RZM
 
 | | Galectin-3C in complex with trifluoroaryltriazole monothiogalactoside derivative:5(Iodine) | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{R})-2-(hydroxymethyl)-6-(3-iodanylphenyl)sulfanyl-4-[4-[3,4,5-tris(fluoranyl)phenyl]-1,2,3-triazol-1-yl]oxane-3,5-diol, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kumar, R, Verteramo, M.L, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2019-06-13 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.345 Å) | | Cite: | Thermodynamic studies of halogen-bond interactions in galectin-3
to be published
|
|
4WBK
 
 | | The 1.37 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with stearic acid | | Descriptor: | Fatty acid-binding protein, heart, STEARIC ACID | | Authors: | Sugiyama, S, Matsuoka, S, Mizohata, E, Matsuoka, D, Murakami, S, Inoue, T, Murata, M. | | Deposit date: | 2014-09-03 | | Release date: | 2015-01-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Molecular Dynamics Simulations of Heart-type Fatty Acid Binding Protein in Apo and Holo Forms, and Hydration Structure Analyses in the Binding Cavity
J.Phys.Chem.B, 119, 2015
|
|
6RZP
 
 | | Multicrystal structure of Proteinase K at room temperature using a multilayer monochromator. | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Sandy, J, Sandy, E, Sanchez-Weatherby, J, Mikolajek, H. | | Deposit date: | 2019-06-13 | | Release date: | 2019-07-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Protein-to-structure pipeline for ambient-temperature crystallography at VMXi
Iucrj, 2023
|
|
