3B72
 
 | | Crystal structure of lysozyme folded in SDS and 2-methyl-2,4-pentanediol | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Michaux, C, Pouyez, J, Wouters, J, Prive, G.G. | | Deposit date: | 2007-10-30 | | Release date: | 2008-09-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Protecting role of cosolvents in protein denaturation by SDS: a structural study.
Bmc Struct.Biol., 8, 2008
|
|
3BH9
 
 | | Crystal Structure of RTY Phosphopeptide Bound to Human Class I MHC HLA-A2 | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Mohammed, F, Cobbold, M, Zarling, A.L, Salim, M, Barrett-Wilt, G.A, Shabanowitz, J, Hunt, D.F, Engelhard, V.H, Willcox, B.E. | | Deposit date: | 2007-11-28 | | Release date: | 2008-10-21 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Phosphorylation-dependent interaction between antigenic peptides and MHC class I: a molecular basis for the presentation of transformed self
Nat.Immunol., 9, 2008
|
|
3BHD
 
 | | Crystal structure of human thiamine triphosphatase (THTPA) | | Descriptor: | CHLORIDE ION, CITRIC ACID, GLYCEROL, ... | | Authors: | Busam, R.D, Lehtio, L, Arrowsmith, C.H, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Herman, M.D, Johansson, A, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Sundstrom, M, Svensson, L, Thorsell, A.G, Tresaugues, L, Van den Berg, S, Weigelt, J, Welin, M, Berglund, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-11-28 | | Release date: | 2007-12-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Human Thiamine Triphosphatase.
To be Published
|
|
3B2S
 
 | | Crystal Structure of F. graminearum TRI101 complexed with Coenzyme A and Deoxynivalenol | | Descriptor: | (3alpha,7alpha)-3,7,15-trihydroxy-12,13-epoxytrichothec-9-en-8-one, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, COENZYME A, ... | | Authors: | Garvey, G.S, Rayment, I. | | Deposit date: | 2007-10-19 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural and Functional Characterization of the TRI101 Trichothecene 3-O-Acetyltransferase from Fusarium sporotrichioides and Fusarium graminearum: KINETIC INSIGHTS TO COMBATING FUSARIUM HEAD BLIGHT
J.Biol.Chem., 283, 2008
|
|
3B9B
 
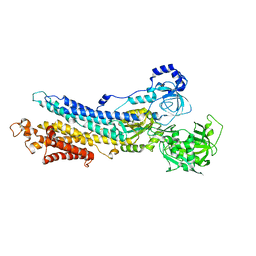 | | Structure of the E2 beryllium fluoride complex of the SERCA Ca2+-ATPase | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Olesen, C, Picard, M, Winther, A.M.L, Morth, J.P, Moller, J.V, Nissen, P. | | Deposit date: | 2007-11-04 | | Release date: | 2007-12-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The structural basis of calcium transport by the calcium pump.
Nature, 450, 2007
|
|
3B76
 
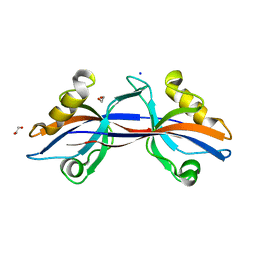 | | Crystal structure of the third PDZ domain of human ligand-of-numb protein-X (LNX1) in complex with the C-terminal peptide from the coxsackievirus and adenovirus receptor | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase LNX, SODIUM ION | | Authors: | Ugochukwu, E, Burgess-Brown, N, Berridge, G, Elkins, J, Bunkoczi, G, Pike, A.C.W, Sundstrom, M, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, Gileadi, O, von Delft, F, Doyle, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-30 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the third PDZ domain of human ligand-of-numb protein-X (LNX1) in complex with the C-terminal peptide from the coxsackievirus and adenovirus receptor.
To be Published
|
|
3B80
 
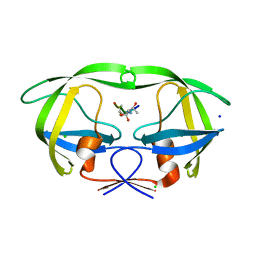 | | HIV-1 protease mutant I54V complexed with gem-diol-amine intermediate NLLTQI | | Descriptor: | CHLORIDE ION, Protease, SODIUM ION, ... | | Authors: | Chumanevich, A.A, Kovalevsky, A.Y, Weber, I.T. | | Deposit date: | 2007-10-31 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Caught in the Act: The 1.5 A Resolution Crystal Structures of the HIV-1 Protease and the I54V Mutant Reveal a Tetrahedral Reaction Intermediate.
Biochemistry, 46, 2007
|
|
3D77
 
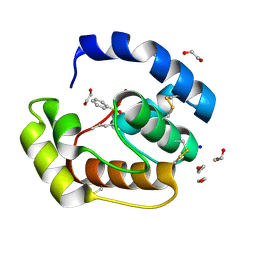 | | Crystal structure of a pheromone binding protein mutant D35N, from Apis mellifera, soaked at pH 4.0 | | Descriptor: | 1,2-ETHANEDIOL, N-BUTYL-BENZENESULFONAMIDE, Pheromone-binding protein ASP1, ... | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-05-20 | | Release date: | 2009-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Queen bee pheromone binding protein pH-induced domain swapping favors pheromone release
J.Mol.Biol., 390, 2009
|
|
3CTZ
 
 | | Structure of human cytosolic X-prolyl aminopeptidase | | Descriptor: | CALCIUM ION, HEXAETHYLENE GLYCOL, MANGANESE (II) ION, ... | | Authors: | Li, X, Lou, Z, Rao, Z. | | Deposit date: | 2008-04-15 | | Release date: | 2008-05-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of human cytosolic X-prolyl aminopeptidase: a double Mn(II)-dependent dimeric enzyme with a novel three-domain subunit
J.Biol.Chem., 283, 2008
|
|
3D1Y
 
 | | Crystal structure of HIV-1 mutant I54V and inhibitor SAQUINA | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, CHLORIDE ION, HIV-1 Protease, ... | | Authors: | Liu, F, Weber, I.T. | | Deposit date: | 2008-05-06 | | Release date: | 2008-05-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Effect of flap mutations on structure of HIV-1 protease and inhibition by saquinavir and darunavir.
J.Mol.Biol., 381, 2008
|
|
2ZKB
 
 | | Urate oxidase complexed with 8-azaxanthine under 2.5 MPa oxygen pressure | | Descriptor: | 8-AZAXANTHINE, OXYGEN MOLECULE, SODIUM ION, ... | | Authors: | Colloc'h, N, Gabison, L, Chiadmi, M, Abraini, J.H, Prange, T. | | Deposit date: | 2008-03-14 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Oxygen pressurized X-ray crystallography: probing the dioxygen binding site in cofactorless urate oxidase and implications for its catalytic mechanism.
Biophys.J., 95, 2008
|
|
3CKU
 
 | |
1Q6X
 
 | | Crystal structure of rat choline acetyltransferase | | Descriptor: | SODIUM ION, choline O-acetyltransferase | | Authors: | Cai, Y, Rodgers, D.W. | | Deposit date: | 2003-08-14 | | Release date: | 2004-06-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Choline acetyltransferase structure reveals distribution of mutations that cause motor disorders.
Embo J., 23, 2004
|
|
3A96
 
 | | Crystal structure of hen egg white lysozyme soaked with 100mM RhCl3 at pH2.2 | | Descriptor: | CHLORIDE ION, Lysozyme C, RHODIUM(III) ION, ... | | Authors: | Abe, S, Koshiyama, T, Ohki, T, Hikage, T, Watanabe, Y, Ueno, T. | | Deposit date: | 2009-10-15 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Elucidation of Metal-Ion Accumulation Induced by Hydrogen Bonds on Protein Surfaces by Using Porous Lysozyme Crystals Containing Rh(III) Ions as the Model Surfaces
Chemistry, 16, 2010
|
|
3AKF
 
 | | Crystal structure of exo-1,5-alpha-L-arabinofuranosidase | | Descriptor: | CHLORIDE ION, GLYCEROL, Putative secreted alpha L-arabinofuranosidase II, ... | | Authors: | Fujimoto, Z, Ichinose, H, Kaneko, S. | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of an Exo-1,5-{alpha}-L-arabinofuranosidase from Streptomyces avermitilis Provides Insights into the Mechanism of Substrate Discrimination between Exo- and Endo-type Enzymes in Glycoside Hydrolase Family 43.
J.Biol.Chem., 285, 2010
|
|
3AHD
 
 | | Phosphoketolase from Bifidobacterium Breve complexed with 2-acetyl-thiamine diphosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-ACETYL-THIAMINE DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Suzuki, R, Katayama, T, Kim, B.-J, Wakagi, T, Shoun, H, Ashida, H, Yamamoto, K, Fushinobu, S. | | Deposit date: | 2010-04-22 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of phosphoketolase: thiamine diphosphate-dependent dehydration mechanism
J.Biol.Chem., 285, 2010
|
|
3A65
 
 | | Crystal structure of 6-aminohexanoate-dimer hydrolase S112A/G181D/H266N mutant with substrate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOIC ACID, 6-aminohexanoate-dimer hydrolase, ... | | Authors: | Kawashima, Y, Shibata, N, Higuchi, Y, Takeo, M, Negoro, S. | | Deposit date: | 2009-08-21 | | Release date: | 2010-09-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Enzymatic Synthesis of Nylon-6 Units in Organic Sol Contained Low-Water: Structural Requirement of 6-Aminohexanoate-Dimer Hydrolase for Efficient Amid Synthesis
To be Published
|
|
3A93
 
 | | Crystal structure of hen egg white lysozyme soaked with 30mM RhCl3 | | Descriptor: | CHLORIDE ION, Lysozyme C, RHODIUM(III) ION, ... | | Authors: | Abe, S, Koshiyama, T, Ohki, T, Hikage, T, Watanabe, Y, Ueno, T. | | Deposit date: | 2009-10-15 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Elucidation of Metal-Ion Accumulation Induced by Hydrogen Bonds on Protein Surfaces by Using Porous Lysozyme Crystals Containing Rh(III) Ions as the Model Surfaces
Chemistry, 16, 2010
|
|
3CPE
 
 | | Crystal Structure of T4 gp17 | | Descriptor: | DNA packaging protein Gp17, PHOSPHATE ION, SODIUM ION | | Authors: | Sun, S, Rossmann, M.G. | | Deposit date: | 2008-03-31 | | Release date: | 2009-01-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structure of the phage T4 DNA packaging motor suggests a mechanism dependent on electrostatic forces
Cell(Cambridge,Mass.), 135, 2008
|
|
3D76
 
 | | Crystal structure of a pheromone binding protein mutant D35N, from Apis mellifera, soaked at pH 7.0 | | Descriptor: | 1,2-ETHANEDIOL, N-BUTYL-BENZENESULFONAMIDE, Pheromone-binding protein ASP1, ... | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-05-20 | | Release date: | 2009-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Queen bee pheromone binding protein pH-induced domain swapping favors pheromone release
J.Mol.Biol., 390, 2009
|
|
1OMO
 
 | | alanine dehydrogenase dimer w/bound NAD (archaeal) | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, alanine dehydrogenase | | Authors: | Gallagher, D.T, Smith, N.N, Holden, M.J, Schroeder, I, Monbouquette, H.G. | | Deposit date: | 2003-02-25 | | Release date: | 2004-07-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structure of alanine dehydrogenase from Archaeoglobus: active site analysis and relation to bacterial cyclodeaminases and mammalian mu crystallin.
J.Mol.Biol., 342, 2004
|
|
7YA4
 
 | |
7YBG
 
 | |
6QZ7
 
 | | Structure of MBP-Mcl-1 in complex with compound 8b | | Descriptor: | (2~{R})-2-[[6-ethyl-5-(1~{H}-indol-4-yl)thieno[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, SODIUM ION, ... | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
6QXJ
 
 | | Structure of MBP-Mcl-1 in complex with compound 6a | | Descriptor: | (2~{R})-2-[[6-ethyl-5-(1~{H}-indol-5-yl)thieno[2,3-d]pyrimidin-4-yl]amino]propanoic acid, Maltose-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, SODIUM ION, ... | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-07 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
