8EEM
 
 | |
8EEH
 
 | |
8EEO
 
 | |
8EEF
 
 | |
8EEN
 
 | |
1OJL
 
 | |
8EEI
 
 | |
1FW6
 
 | | CRYSTAL STRUCTURE OF A TAQ MUTS-DNA-ADP TERNARY COMPLEX | | Descriptor: | 5'-D(*GP*CP*GP*AP*CP*GP*CP*TP*AP*GP*CP*GP*TP*GP*CP*GP*GP*CP*TP*CP*GP*TP*C)-3', 5'-D(*GP*GP*AP*CP*GP*AP*GP*CP*CP*GP*CP*CP*GP*CP*TP*AP*GP*CP*GP*TP*CP*G)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Junop, M.S, Obmolova, G, Rausch, K, Hsieh, P, Yang, W. | | Deposit date: | 2000-09-21 | | Release date: | 2001-02-19 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Composite active site of an ABC ATPase: MutS uses ATP to verify mismatch recognition and authorize DNA repair.
Mol.Cell, 7, 2001
|
|
3KLL
 
 | | Crystal structure of Lactobacillus reuteri N-terminally truncated glucansucrase GTF180-maltose complex | | Descriptor: | CALCIUM ION, GLYCEROL, Glucansucrase, ... | | Authors: | Vujicic-Zagar, A, Pijning, T, Kralj, S, Eeuwema, W, Dijkhuizen, L, Dijkstra, B.W. | | Deposit date: | 2009-11-08 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a 117 kDa glucansucrase fragment provides insight into evolution and product specificity of GH70 enzymes
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
8EEJ
 
 | |
8EEK
 
 | |
8EEL
 
 | |
2WMM
 
 | | Crystal structure of the hinge domain of MukB | | Descriptor: | Chromosome partition protein MukB, D-MALATE | | Authors: | Ku, B, Oh, B.-H. | | Deposit date: | 2009-07-01 | | Release date: | 2010-01-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the MukB hinge domain with coiled-coil stretches and its functional implications.
Proteins, 78, 2010
|
|
1FUO
 
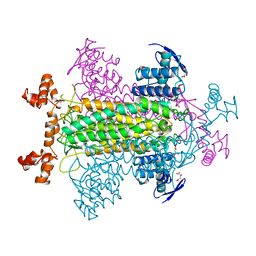 | | FUMARASE C WITH BOUND CITRATE | | Descriptor: | CITRIC ACID, D-MALATE, FUMARASE C | | Authors: | Weaver, T, Banaszak, L. | | Deposit date: | 1996-08-29 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystallographic studies of the catalytic and a second site in fumarase C from Escherichia coli.
Biochemistry, 35, 1996
|
|
1Q51
 
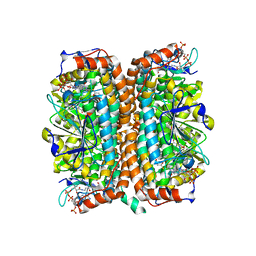 | | Crystal Structure of Mycobacterium tuberculosis MenB in Complex with Acetoacetyl-Coenzyme A, a Key Enzyme in Vitamin K2 Biosynthesis | | Descriptor: | ACETOACETYL-COENZYME A, menB | | Authors: | Truglio, J.J, Theis, K, Feng, Y, Gajda, R, Machutta, C, Tonge, P.J, Kisker, C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-08-05 | | Release date: | 2004-01-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis MenB, a key enzyme in vitamin K2 biosynthesis.
J.Biol.Chem., 278, 2003
|
|
1ASU
 
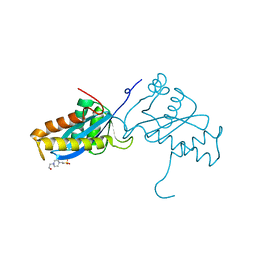 | | AVIAN SARCOMA VIRUS INTEGRASE CATALYTIC CORE DOMAIN CRYSTALLIZED FROM 2% PEG 400, 2M AMMONIUM SULFATE, HEPES PH 7.5 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AVIAN SARCOMA VIRUS INTEGRASE | | Authors: | Bujacz, G, Jaskolski, M, Alexandratos, J, Wlodawer, A. | | Deposit date: | 1995-08-25 | | Release date: | 1995-11-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-resolution structure of the catalytic domain of avian sarcoma virus integrase.
J.Mol.Biol., 253, 1995
|
|
3JS6
 
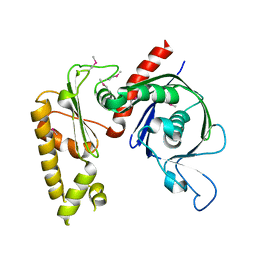 | | Crystal structure of apo psk41 parM protein | | Descriptor: | Uncharacterized ParM protein | | Authors: | Schumacher, M.A, Xu, W, Firth, N. | | Deposit date: | 2009-09-09 | | Release date: | 2010-01-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and filament dynamics of the pSK41 actin-like ParM protein: implications for plasmid DNA segregation.
J.Biol.Chem., 285, 2010
|
|
5Z36
 
 | | An anthrahydroquino-Gama-pyrone synthase Txn09 complexed with PDM | | Descriptor: | 11-hydroxy-2-[(2S)-2-hydroxybutan-2-yl]-5-methyl-4H-anthra[1,2-b]pyran-4,7,12-trione, TxnO9 | | Authors: | Song, Y.J, Cao, C.Y. | | Deposit date: | 2018-01-05 | | Release date: | 2018-12-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Enzymology of Anthraquinone-gamma-Pyrone Ring Formation in Complex Aromatic Polyketide Biosynthesis.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5WK0
 
 | |
3N6W
 
 | | Crystal structure of human gamma-butyrobetaine hydroxylase | | Descriptor: | Gamma-butyrobetaine dioxygenase, SULFATE ION, ZINC ION | | Authors: | Rumnieks, J, Zeltins, A, Leonchiks, A, Kazaks, A, Kotelovica, S, Tars, K. | | Deposit date: | 2010-05-26 | | Release date: | 2010-07-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of human gamma-butyrobetaine hydroxylase.
Biochem.Biophys.Res.Commun., 398, 2010
|
|
4NDV
 
 | |
3K52
 
 | |
1G6E
 
 | | ANTIFUNGAL PROTEIN FROM STREPTOMYCES TENDAE TU901, 30-CONFORMERS ENSEMBLE | | Descriptor: | ANTIFUNGAL PROTEIN | | Authors: | Campos-Olivas, R, Bormann, C, Hoerr, I, Jung, G, Gronenborn, A.M. | | Deposit date: | 2000-11-04 | | Release date: | 2001-03-28 | | Last modified: | 2022-12-21 | | Method: | SOLUTION NMR | | Cite: | Solution structure, backbone dynamics and chitin binding of the anti-fungal protein from Streptomyces tendae TU901.
J.Mol.Biol., 308, 2001
|
|
6I7K
 
 | | Crystal structure of monomeric FICD mutant L258D complexed with MgATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Adenosine monophosphate-protein transferase FICD, ETHANOL, ... | | Authors: | Perera, L.A, Yan, Y, Read, R.J, Ron, D. | | Deposit date: | 2018-11-16 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | An oligomeric state-dependent switch in the ER enzyme FICD regulates AMPylation and deAMPylation of BiP.
Embo J., 38, 2019
|
|
3KQH
 
 | |
