5M6H
 
 | | Small Molecule inhibitors of IAP | | Descriptor: | 1-[3,3-dimethyl-6-(phenylmethyl)-2~{H}-pyrrolo[3,2-b]pyridin-1-yl]-2-[(2~{R},5~{R})-5-methyl-2-morpholin-4-ylcarbonyl-piperazin-4-ium-1-yl]ethanone, E3 ubiquitin-protein ligase XIAP, SODIUM ION, ... | | Authors: | Williams, P.A. | | Deposit date: | 2016-10-25 | | Release date: | 2017-05-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of a Potent Nonpeptidomimetic, Small-Molecule Antagonist of Cellular Inhibitor of Apoptosis Protein 1 (cIAP1) and X-Linked Inhibitor of Apoptosis Protein (XIAP).
J. Med. Chem., 60, 2017
|
|
6I49
 
 | | Structure of P. aeruginosa LpxC with compound 17a: (2R)-N-Hydroxy-2-methyl-2-(methylsulfonyl)-4(6((4(morpholinomethyl)phenyl)ethynyl)-3-oxo-1H-pyrrolo[1,2-c]imidazol-2(3H)yl)butanamide | | Descriptor: | (2~{R})-2-methyl-2-methylsulfonyl-4-[6-[2-[4-(morpholin-4-ylmethyl)phenyl]ethynyl]-3-oxidanylidene-1~{H}-pyrrolo[1,2-c]imidazol-2-yl]-~{N}-oxidanyl-butanamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Surivet, J.-P, Panchaud, P, Specklin, J.-L, Diethelm, S, Blumstein, A.-C, Gauvin, J.-C, Jacob, L, Masse, F, Mathieu, G, Mirre, A, Schmitt, C, Enderlin-Paput, M, Lange, R, Bur, D, Tidten-Luksch, N, Gnerre, C, Seeland, S, Hermann, C, Locher, H.H, Seiler, P, Mac Sweeney, A, Hubschwerlen, C, Ritz, D, Rueedi, G. | | Deposit date: | 2018-11-09 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Discovery of Novel Inhibitors of LpxC Displaying Potent in Vitro Activity against Gram-Negative Bacteria.
J.Med.Chem., 63, 2020
|
|
7BZ4
 
 | |
6I6C
 
 | | SEPIAPTERIN REDUCTASE IN COMPLEX WITH COMPOUND 2 | | Descriptor: | (1~{R},2~{S},4~{S})-~{N}-(3-chloranyl-4-cyano-phenyl)sulfonylbicyclo[2.2.1]heptane-2-carboxamide, 1,2-ETHANEDIOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Alen, J, Schade, M, Wagener, M, Blaesse, M. | | Deposit date: | 2018-11-15 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Fragment-Based Discovery of Novel Potent Sepiapterin Reductase Inhibitors.
J.Med.Chem., 62, 2019
|
|
6I17
 
 | | CRYSTAL STRUCTURE OF FASCIN IN COMPLEX WITH COMPOUND 24 | | Descriptor: | 1,2-ETHANEDIOL, 2-[(3,4-dichlorophenyl)methyl]-~{N}-(1-methylpyrazol-4-yl)-1-oxidanylidene-6-piperidin-4-yl-2,7-naphthyridine-4-carboxamide, ACETATE ION, ... | | Authors: | Schuettelkopf, A.W. | | Deposit date: | 2018-10-27 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structure-based design, synthesis and biological evaluation of a novel series of isoquinolone and pyrazolo[4,3-c]pyridine inhibitors of fascin 1 as potential anti-metastatic agents.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6ZN6
 
 | | Protein polybromo-1 (PB1 BD2) Bound To MW278 | | Descriptor: | 1,2-ETHANEDIOL, 2-[6-azanyl-5-[(3~{R})-3-phenoxypiperidin-1-yl]pyridazin-3-yl]phenol, Protein polybromo-1 | | Authors: | Preuss, F, Mathea, S, Chatterjee, D, Wanior, M, Joerger, A.C, Knapp, S. | | Deposit date: | 2020-07-06 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Pan-SMARCA/PB1 Bromodomain Inhibitors and Their Role in Regulating Adipogenesis.
J.Med.Chem., 63, 2020
|
|
5OFU
 
 | | Crystal structure of Leishmania major fructose-1,6-bisphosphatase in T-state. | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Yuan, M, Vasquez-Valdivieso, M.G, McNae, I.W, Michels, P.A.M, Fothergill-Gilmore, L.A, Walkinshaw, M.D. | | Deposit date: | 2017-07-11 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structures of Leishmania Fructose-1,6-Bisphosphatase Reveal Species-Specific Differences in the Mechanism of Allosteric Inhibition.
J. Mol. Biol., 429, 2017
|
|
9EB4
 
 | | Chicken YF1.7*1 presenting N-palmitoylated glycine | | Descriptor: | Beta-2-microglobulin, CHLORIDE ION, GLYCEROL, ... | | Authors: | Khandokar, Y, Wang, C.J, Rossjohn, J, Le Nours, J. | | Deposit date: | 2024-11-11 | | Release date: | 2025-07-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular basis for presentation of N-myristoylated peptides by the chicken YF1∗7.1 molecule.
J.Biol.Chem., 301, 2025
|
|
9EB3
 
 | | Chicken YF1.7*1 presenting N-myristoylated glycine | | Descriptor: | Beta-2-microglobulin, CHLORIDE ION, GLYCEROL, ... | | Authors: | Khandokar, Y, Wang, C.J.H, Rossjohn, J, Le Nours, J. | | Deposit date: | 2024-11-11 | | Release date: | 2025-07-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis for presentation of N-myristoylated peptides by the chicken YF1∗7.1 molecule.
J.Biol.Chem., 301, 2025
|
|
5NYL
 
 | |
5Z5H
 
 | | Crystal structure of a thermostable glycoside hydrolase family 43 {beta}-1,4-xylosidase from Geobacillus thermoleovorans IT-08 in complex with D-xylose | | Descriptor: | Beta-xylosidase, CALCIUM ION, alpha-D-xylopyranose | | Authors: | Rohman, A, van Oosterwijk, N, Puspaningsih, N.N.T, Dijkstra, B.W. | | Deposit date: | 2018-01-18 | | Release date: | 2018-04-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of product inhibition by arabinose and xylose of the thermostable GH43 beta-1,4-xylosidase from Geobacillus thermoleovorans IT-08.
PLoS ONE, 13, 2018
|
|
8B8D
 
 | | multimerization domain of Gaboon Viper Virus 1 | | Descriptor: | Phosphoprotein | | Authors: | Tarbouriech, N, Legrand, P, Bouhris, J.M, Horie, M, Tomonaga, K, Crepin, T. | | Deposit date: | 2022-10-04 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Borna Disease Virus 1 Phosphoprotein Forms a Tetramer and Interacts with Host Factors Involved in DNA Double-Strand Break Repair and mRNA Processing.
Viruses, 14, 2022
|
|
5KMO
 
 | | TrkA JM-kinase with 1-(2-methyl-4-phenyl-pyrimidin-5-yl)-3-(2-pyridyl)urea | | Descriptor: | 1-(2-methyl-4-phenyl-pyrimidin-5-yl)-3-pyridin-2-yl-urea, High affinity nerve growth factor receptor | | Authors: | Su, H.P. | | Deposit date: | 2016-06-27 | | Release date: | 2016-12-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Structural characterization of nonactive site, TrkA-selective kinase inhibitors.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7CBQ
 
 | | Crystal structure of PDE4D catalytic domain in complex with Apremilast | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, N-{2-[(1S)-1-(3-ethoxy-4-methoxyphenyl)-2-(methylsulfonyl)ethyl]-1,3-dioxo-2,3-dihydro-1H-isoindol-4-yl}acetamide, ... | | Authors: | Zhang, X.L, Xu, Y.C. | | Deposit date: | 2020-06-13 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Design, synthesis, and biological evaluation of tetrahydroisoquinolines derivatives as novel, selective PDE4 inhibitors for antipsoriasis treatment.
Eur.J.Med.Chem., 211, 2021
|
|
1AI1
 
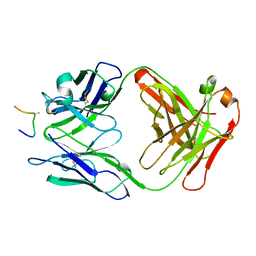 | | HIV-1 V3 LOOP MIMIC | | Descriptor: | AIB142, IGG1-KAPPA 59.1 FAB (HEAVY CHAIN), IGG1-KAPPA 59.1 FAB (LIGHT CHAIN) | | Authors: | Ghiara, J.B, Wilson, I.A. | | Deposit date: | 1996-11-06 | | Release date: | 1997-05-15 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based design of a constrained peptide mimic of the HIV-1 V3 loop neutralization site.
J.Mol.Biol., 266, 1997
|
|
9EU2
 
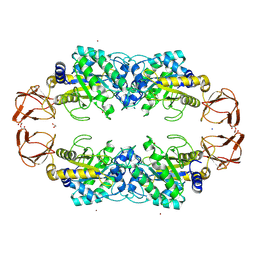 | | GH29A alpha-L-fucosidase | | Descriptor: | 1,2-ETHANEDIOL, Alpha-L-fucosidase, SODIUM ION, ... | | Authors: | Yang, Y.Y, Zeuner, B, Morth, J.P. | | Deposit date: | 2024-03-27 | | Release date: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural elucidation and characterization of GH29A alpha-l-fucosidases and the effect of pH on their transglycosylation.
Febs J., 292, 2025
|
|
6GK8
 
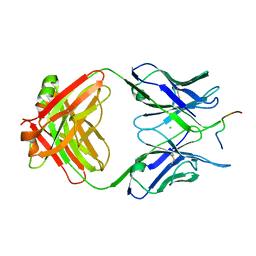 | | Crystal structure of anti-tau antibody dmCBTAU-28.1, double mutant (S32R, E35K) of CBTAU-28.1, in complex with Tau peptide A7731 (residues 52-71) | | Descriptor: | CHLORIDE ION, HUMAN FAB ANTIBODY FRAGMENT OF CBTAU-28.1(S32R;E35K), TAU PEPTIDE A7731 (RESIDUES 52-71) | | Authors: | Steinbacher, S, Mrosek, M, Juraszek, J. | | Deposit date: | 2018-05-18 | | Release date: | 2018-06-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A common antigenic motif recognized by naturally occurring human VH5-51/VL4-1 anti-tau antibodies with distinct functionalities.
Acta Neuropathol Commun, 6, 2018
|
|
7CX9
 
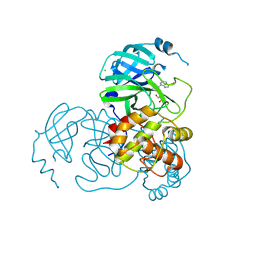 | | Crystal structure of the SARS-CoV-2 main protease in complex with INZ-1 | | Descriptor: | 3-iodanyl-1~{H}-indazole-7-carbaldehyde, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Zeng, R, Liu, X.L, Qiao, J.X, Nan, J.S, Wang, Y.F, Li, Y.S, Yang, S.Y, Lei, J. | | Deposit date: | 2020-09-01 | | Release date: | 2020-09-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of the SARS-CoV-2 main protease in complex with INZ-1
To Be Published
|
|
7NRG
 
 | |
9ENW
 
 | |
5EXJ
 
 | | Crystal structure of M. tuberculosis lipoyl synthase at 1.64 A resolution | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, IRON/SULFUR CLUSTER, Lipoyl synthase | | Authors: | McLaughlin, M.I, Lanz, N.D, Goldman, P.J, Lee, K.-H, Booker, S.J, Drennan, C.L. | | Deposit date: | 2015-11-23 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystallographic snapshots of sulfur insertion by lipoyl synthase.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7CTS
 
 | | Open form of PET-degrading cutinase Cut190 with thermostability-improving mutations of S226P/R228S/Q138A/D250C-E296C/Q123H/N202H and S176A inactivation | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Alpha/beta hydrolase family protein, BICINE, ... | | Authors: | Emori, M, Numoto, N, Senga, A, Bekker, G.J, Kamiya, N, Ito, N, Kawai, F, Oda, M. | | Deposit date: | 2020-08-20 | | Release date: | 2021-02-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural basis of mutants of PET-degrading enzyme from Saccharomonospora viridis AHK190 with high activity and thermal stability.
Proteins, 89, 2021
|
|
7CKA
 
 | |
7N9X
 
 | | CA-targeting nanobody is a tool for studying HIV-1 capsid lattice interactions | | Descriptor: | Capsid protein, Nanobody, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Gerber, E.E, Digianantonio, K.M, Tripler, T.N, Smaga, S.S, Summers, B.J, Xiong, Y. | | Deposit date: | 2021-06-18 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.511 Å) | | Cite: | CA-targeting nanobody is a tool for studying HIV-1 capsid lattice interactions
To Be Published
|
|
7NU6
 
 | | Crystal Structure of Neisseria gonorrhoeae LeuRS in Complex with ATP in Conformation 1 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Leucine--tRNA ligase, ... | | Authors: | Pang, L, Strelkov, S.V, Weeks, S.D. | | Deposit date: | 2021-03-11 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Partitioning of the initial catalytic steps of leucyl-tRNA synthetase is driven by an active site peptide-plane flip.
Commun Biol, 5, 2022
|
|
