4LFK
 
 | |
5YCP
 
 | | Human PPARgamma ligand binding domain complexed with Rosiglitazone | | Descriptor: | 2,4-THIAZOLIDIINEDIONE, 5-[[4-[2-(METHYL-2-PYRIDINYLAMINO)ETHOXY]PHENYL]METHYL]-(9CL), Nuclear receptor coactivator 1, ... | | Authors: | Jang, J.Y, Han, B.W. | | Deposit date: | 2017-09-07 | | Release date: | 2018-09-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Enhanced Anti-Diabetic Efficacy of Lobeglitazone on PPAR gamma.
Sci Rep, 8, 2018
|
|
4LFL
 
 | |
3ICT
 
 | | Crystal structure of reduced Bacillus anthracis CoADR-RHD | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, COENZYME A, Coenzyme A-Disulfide Reductase, ... | | Authors: | Wallen, J.R, Claiborne, A. | | Deposit date: | 2009-07-18 | | Release date: | 2009-11-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and catalytic properties of Bacillus anthracis CoADR-RHD: implications for flavin-linked sulfur trafficking.
Biochemistry, 48, 2009
|
|
3ITC
 
 | | Crystal structure of Sco3058 with bound citrate and glycerol | | Descriptor: | CITRIC ACID, GLYCEROL, ZINC ION, ... | | Authors: | Nguyen, T.T, Cummings, J.A, Tsai, C.-L, Barondeau, D.P, Raushel, F.M. | | Deposit date: | 2009-08-28 | | Release date: | 2010-02-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure, mechanism, and substrate profile for Sco3058: the closest bacterial homologue to human renal dipeptidase
Biochemistry, 49, 2010
|
|
5YQJ
 
 | | Crystal structure of the first StARkin domain of Lam4 | | Descriptor: | Membrane-anchored lipid-binding protein LAM4 | | Authors: | Im, Y.J, Tong, J.S. | | Deposit date: | 2017-11-06 | | Release date: | 2018-01-31 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of sterol recognition and nonvesicular transport by lipid transfer proteins anchored at membrane contact sites
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5YCQ
 
 | | Unique Specificity-Enhancing Factor for the AAA+ Lon Protease | | Descriptor: | Heat shock protein HspQ | | Authors: | Abe, Y, Shioi, S, Kita, S, Nakata, H, Maenaka, K, Kohda, D, Katayama, T, Ueda, T. | | Deposit date: | 2017-09-08 | | Release date: | 2018-04-11 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | X-ray crystal structure of Escherichia coli HspQ, a protein involved in the retardation of replication initiation
FEBS Lett., 591, 2017
|
|
7QHB
 
 | | Active state of GluA1/2 in complex with TARP gamma 8, L-glutamate and CTZ | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (7Z)-hexadec-7-enoate, CYCLOTHIAZIDE, ... | | Authors: | Herguedas, B, Kohegyi, B, Zhang, D, Greger, I.H. | | Deposit date: | 2021-12-11 | | Release date: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mechanisms underlying TARP modulation of the GluA1/2-gamma 8 AMPA receptor.
Nat Commun, 13, 2022
|
|
7QHH
 
 | | Desensitized state of GluA1/2 AMPA receptor in complex with TARP-gamma 8 (TMD-LBD) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (7Z)-hexadec-7-enoate, GLUTAMIC ACID, ... | | Authors: | Herguedas, B, Kohegyi, B, Dohrke, J.N, Watson, J.F, Zhang, D, Ho, H, Shaikh, S, Lape, R, Krieger, J.M, Greger, I.H. | | Deposit date: | 2021-12-12 | | Release date: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Mechanisms underlying TARP modulation of the GluA1/2-gamma 8 AMPA receptor.
Nat Commun, 13, 2022
|
|
5YQQ
 
 | |
4LFN
 
 | |
3KLK
 
 | | Crystal structure of Lactobacillus reuteri N-terminally truncated glucansucrase GTF180 in triclinic apo- form | | Descriptor: | CALCIUM ION, GLYCEROL, Glucansucrase | | Authors: | Vujicic-Zagar, A, Pijning, T, Kralj, S, Eeuwema, W, Dijkhuizen, L, Dijkstra, B.W. | | Deposit date: | 2009-11-08 | | Release date: | 2010-11-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of a 117 kDa glucansucrase fragment provides insight into evolution and product specificity of GH70 enzymes
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
8T4M
 
 | | Closed human HCN1 F186C S264C bound to cAMP, reconstituted in LMNG + SPL | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 | | Authors: | Burtscher, V, Mount, J, Cowgill, J, Chang, Y, Bickel, K, Yuan, P, Chanda, B. | | Deposit date: | 2023-06-09 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structural basis for hyperpolarization-dependent opening of human HCN1 channel.
Nat Commun, 15, 2024
|
|
5K7L
 
 | |
8T1D
 
 | | Open-state cryo-EM structure of full-length human TRPV4 in complex with agonist 4a-PDD | | Descriptor: | (1aR,1bS,4aS,7aS,7bS,8R,9R,9aS)-9a-(decanoyloxy)-4a,7b-dihydroxy-3-(hydroxymethyl)-1,1,6,8-tetramethyl-5-oxo-1a,1b,4,4a,5,7a,7b,8,9,9a-decahydro-1H-cyclopropa[3,4]benzo[1,2-e]azulen-9-yl decanoate, Transient receptor potential cation channel subfamily V member 4/Enhanced green fluorescent protein chimera | | Authors: | Talyzina, I.A, Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2023-06-02 | | Release date: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structure of human TRPV4 in complex with GTPase RhoA.
Nat Commun, 14, 2023
|
|
7KFO
 
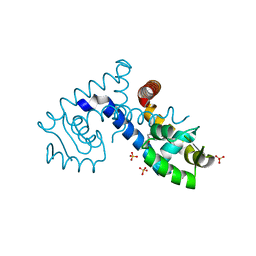 | | Crystal structure of the MarR family transcriptional regulator from Variovorax paradoxus bound to Indole 3 acetic acid | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, SULFATE ION, Transcriptional regulator, ... | | Authors: | Walton, W.G, Redinbo, M.R, Dangl, J.L. | | Deposit date: | 2020-10-14 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Diverse MarR bacterial regulators of auxin catabolism in the plant microbiome.
Nat Microbiol, 7, 2022
|
|
8T1F
 
 | | Cryo-EM structure of full-length human TRPV4 in complex with antagonist HC-067047 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-methyl-1-[3-(morpholin-4-yl)propyl]-5-phenyl-N-[3-(trifluoromethyl)phenyl]-1H-pyrrole-3-carboxamide, Transient receptor potential cation channel subfamily V member 4/Enhanced green fluorescent protein chimera | | Authors: | Talyzina, I.A, Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2023-06-02 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structure of human TRPV4 in complex with GTPase RhoA.
Nat Commun, 14, 2023
|
|
8T1B
 
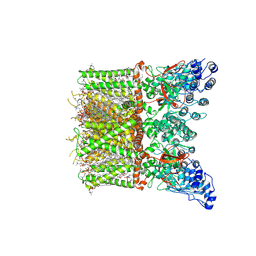 | | Cryo-EM structure of full-length human TRPV4 in apo state | | Descriptor: | (2R)-2-{[(4-O-hexopyranosyl-beta-D-glucopyranosyl)oxy]methyl}-4-{[(25R)-5beta,14beta,17beta-spirostan-3beta-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, SODIUM ION, Transient receptor potential cation channel subfamily V member 4/Enhanced green fluorescent protein chimera, ... | | Authors: | Nadezhdin, K.D, Talyzina, I.A, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2023-06-02 | | Release date: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of human TRPV4 in complex with GTPase RhoA.
Nat Commun, 14, 2023
|
|
7KFQ
 
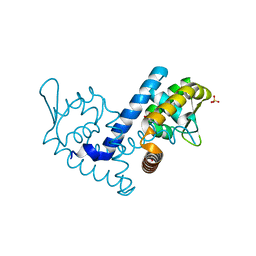 | | Crystal structure of the MarR family transcriptional regulator from Variovorax paradoxus bound to 1H-Indole-3-Carboxylic Acid | | Descriptor: | 1H-INDOLE-3-CARBOXYLIC ACID, SULFATE ION, Transcriptional regulator, ... | | Authors: | Walton, W.G, Redinbo, M.R, Dangl, J.L. | | Deposit date: | 2020-10-14 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Diverse MarR bacterial regulators of auxin catabolism in the plant microbiome.
Nat Microbiol, 7, 2022
|
|
7KFS
 
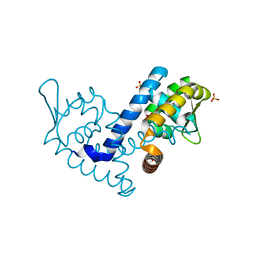 | | Crystal structure of the MarR family transcriptional regulator from Variovorax paradoxus bound to Benzoic Acid | | Descriptor: | BENZOIC ACID, SULFATE ION, Transcriptional regulator, ... | | Authors: | Walton, W.G, Redinbo, M.R, Dangl, J.L. | | Deposit date: | 2020-10-14 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Diverse MarR bacterial regulators of auxin catabolism in the plant microbiome.
Nat Microbiol, 7, 2022
|
|
8T1E
 
 | | Closed-state cryo-EM structure of full-length human TRPV4 in the presence of 4a-PDD | | Descriptor: | (2R)-2-{[(4-O-hexopyranosyl-beta-D-glucopyranosyl)oxy]methyl}-4-{[(25R)-5beta,14beta,17beta-spirostan-3beta-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, SODIUM ION, Transient receptor potential cation channel subfamily V member 4,Enhanced green fluorescent protein, ... | | Authors: | Talyzina, I.A, Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2023-06-02 | | Release date: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structure of human TRPV4 in complex with GTPase RhoA.
Nat Commun, 14, 2023
|
|
8T1C
 
 | | Cryo-EM structure of human TRPV4 ankyrin repeat domain in complex with GTPase RhoA | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Transforming protein RhoA, Transient receptor potential cation channel subfamily V member 4-Enhanced green fluorescent protein chimera | | Authors: | Nadezhdin, K.D, Talyzina, I.A, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2023-06-02 | | Release date: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structure of human TRPV4 in complex with GTPase RhoA.
Nat Commun, 14, 2023
|
|
1CCZ
 
 | | CRYSTAL STRUCTURE OF THE CD2-BINDING DOMAIN OF CD58 (LYMPHOCYTE FUNCTION-ASSOCIATED ANTIGEN 3) AT 1.8-A RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (CD58) | | Authors: | Ikemizu, S, Sparks, L.M, Van Der Merwe, P.A, Harlos, K, Stuart, D.I, Jones, E.Y, Davis, S.J. | | Deposit date: | 1999-03-02 | | Release date: | 1999-04-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the CD2-binding domain of CD58 (lymphocyte function-associated antigen 3) at 1.8-A resolution.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
7KJQ
 
 | | Crystal structure of the MarR family transcriptional regulator from Variovorax paradoxus bound to Picloram | | Descriptor: | Picloram, SULFATE ION, Transcriptional regulator, ... | | Authors: | Walton, W.G, Redinbo, M.R, Dangl, J.L. | | Deposit date: | 2020-10-26 | | Release date: | 2021-12-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Diverse MarR bacterial regulators of auxin catabolism in the plant microbiome.
Nat Microbiol, 7, 2022
|
|
7KH3
 
 | | Crystal structure of the MarR family transcriptional regulator from Variovorax paradoxus bound to Indole 3 propionic acid | | Descriptor: | INDOLYLPROPIONIC ACID, SULFATE ION, Transcriptional regulator, ... | | Authors: | Walton, W.G, Redinbo, M.R, Dangl, J.L. | | Deposit date: | 2020-10-19 | | Release date: | 2021-12-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Diverse MarR bacterial regulators of auxin catabolism in the plant microbiome.
Nat Microbiol, 7, 2022
|
|
