7AQS
 
 | | Crystal structure of E. coli DPS in space group P1 | | Descriptor: | DNA protection during starvation protein, FE (III) ION | | Authors: | Jakob, R.P, Pipercevic, J, Righetto, R, Goldie, K, Stahlberg, H, Maier, T, Hiller, S. | | Deposit date: | 2020-10-22 | | Release date: | 2021-09-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification of a Dps contamination in Mitomycin-C-induced expression of Colicin Ia.
Biochim Biophys Acta Biomembr, 1863, 2021
|
|
1Q7C
 
 | | The structure of betaketoacyl-[ACP] reductase Y151F mutant in complex with NADPH fragment | | Descriptor: | 3-oxoacyl-[acyl-carrier protein] reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Price, A.C, Zhang, Y.-M, Rock, C.O, White, S.M. | | Deposit date: | 2003-08-17 | | Release date: | 2004-02-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Cofactor-Induced Conformational Rearrangements Establish a Catalytically Competent Active Site and a
Proton Relay Conduit in FabG
Structure, 12, 2004
|
|
7AWI
 
 | | Crystal structure of human butyrylcholinesterase in complex with tert-butyl 3-(((2-((1-benzyl-1H-indol-4-yl)oxy)ethyl)amino)methyl]piperidine-1-carboxylate | | Descriptor: | 2-((1-benzyl-1H-indol-4-yl)oxy)-N-((1-(tert-butoxycarbonyl)piperidin-3-yl)methyl)ethan-1-aminium, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brazzolotto, X, Goral, I, Wieckowska, A. | | Deposit date: | 2020-11-08 | | Release date: | 2021-09-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Development and crystallography-aided SAR studies of multifunctional BuChE inhibitors and 5-HT 6 R antagonists with beta-amyloid anti-aggregation properties.
Eur.J.Med.Chem., 225, 2021
|
|
1Q7T
 
 | | Rv1170 (MshB) from Mycobacterium tuberculosis | | Descriptor: | SULFATE ION, hypothetical protein Rv1170, octyl beta-D-glucopyranoside | | Authors: | McCarthy, A.A, Peterson, N.A, Knijff, R, Baker, E.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-08-20 | | Release date: | 2004-01-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of MshB from Mycobacterium tuberculosis, a Deacetylase Involved in Mycothiol Biosynthesis.
J.Mol.Biol., 335, 2004
|
|
7AWG
 
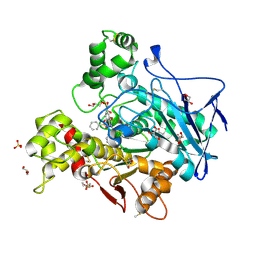 | | Crystal structure of human butyrylcholinesterase in complex with (2-((1-(benzenesulfonyl)-1H-indol-4-yl)oxy)ethyl)(benzyl)amine | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brazzolotto, X, Wichur, T, Wieckowska, A. | | Deposit date: | 2020-11-08 | | Release date: | 2021-09-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development and crystallography-aided SAR studies of multifunctional BuChE inhibitors and 5-HT 6 R antagonists with beta-amyloid anti-aggregation properties.
Eur.J.Med.Chem., 225, 2021
|
|
7AMV
 
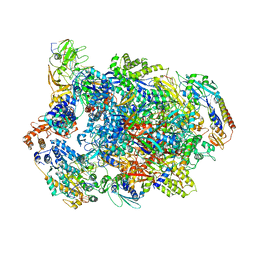 | | Atomic structure of the poxvirus transcription pre-initiation complex in the initially melted state | | Descriptor: | ATP-dependent helicase VETFS, DNA-directed RNA polymerase, DNA-directed RNA polymerase 147 kDa polypeptide, ... | | Authors: | Grimm, C, Bartuli, J, Fischer, U. | | Deposit date: | 2020-10-09 | | Release date: | 2021-10-06 | | Last modified: | 2021-10-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of the complete poxvirus transcription initiation process.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7AWH
 
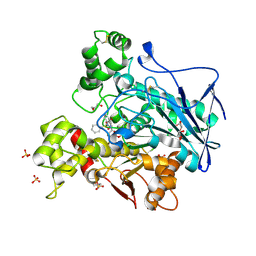 | | Crystal structure of human butyrylcholinesterase in complex with tert-butyl 3-(((2-((1-(benzenesulfonyl)-1H-indol-4-yl)oxy)ethyl)amino)methyl)piperidine-1-carboxylate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brazzolotto, X, Wichur, T, Wieckowska, A. | | Deposit date: | 2020-11-08 | | Release date: | 2021-09-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Development and crystallography-aided SAR studies of multifunctional BuChE inhibitors and 5-HT 6 R antagonists with beta-amyloid anti-aggregation properties.
Eur.J.Med.Chem., 225, 2021
|
|
7B15
 
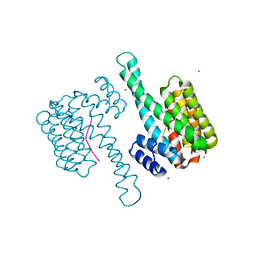 | |
7B13
 
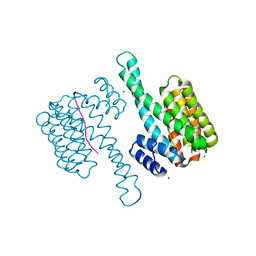 | |
7B1O
 
 | | Crystal structure of the indoleamine 2,3-dioxygenase 1 (IDO1) in complex with compound 22 | | Descriptor: | 4-chloranyl-N-[(1R)-1-[(1S,5R)-3-quinolin-4-yloxy-6-bicyclo[3.1.0]hexanyl]propyl]benzamide, Indoleamine 2,3-dioxygenase 1 | | Authors: | Lammens, A, Krapp, S, Lewis, R.T, Hamilton, M.M. | | Deposit date: | 2020-11-25 | | Release date: | 2021-09-29 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Discovery of IACS-9779 and IACS-70465 as Potent Inhibitors Targeting Indoleamine 2,3-Dioxygenase 1 (IDO1) Apoenzyme.
J.Med.Chem., 64, 2021
|
|
7ATH
 
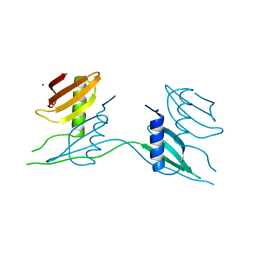 | | Crystal structure of UipA | | Descriptor: | UipA, ZINC ION | | Authors: | Bremond, N, Gallois, N, Legrand, P, Chapon, V, Arnoux, P. | | Deposit date: | 2020-10-30 | | Release date: | 2021-10-06 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.343 Å) | | Cite: | Discovery and characterization of UipA, a uranium- and iron-binding PepSY protein involved in uranium tolerance by soil bacteria.
Isme J, 16, 2022
|
|
1QJA
 
 | | 14-3-3 ZETA/PHOSPHOPEPTIDE COMPLEX (MODE 2) | | Descriptor: | 14-3-3 PROTEIN ZETA, PHOSPHOPEPTIDE | | Authors: | Rittinger, K, Budman, J, Xu, J, Volinia, S, Cantley, L.C, Smerdon, S.J, Gamblin, S.J, Yaffe, M.B. | | Deposit date: | 1999-06-23 | | Release date: | 1999-09-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of 14-3-3 Phosphopeptide Complexes Identifies a Dual Role for the Nuclear Export Signal of 14-3-3 in Ligand Binding
Mol.Cell, 4, 1999
|
|
1QE3
 
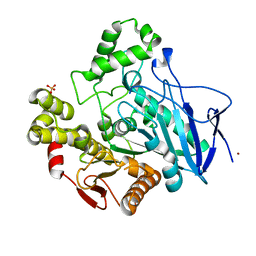 | | PNB ESTERASE | | Descriptor: | PARA-NITROBENZYL ESTERASE, SULFATE ION, ZINC ION | | Authors: | Spiller, B, Gershenson, A, Arnold, F, Stevens, R. | | Deposit date: | 1999-07-12 | | Release date: | 1999-07-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A structural view of evolutionary divergence.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
5BUM
 
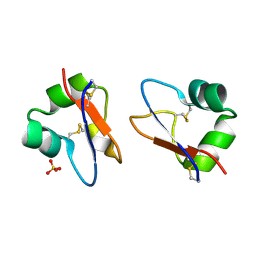 | | Crystal Structure of LysM domain from Equisetum arvense chitinase A | | Descriptor: | Chitinase A, SULFATE ION | | Authors: | Kitaoku, Y, Numata, T, Ohnuma, T, Taira, T, Fukamizo, T. | | Deposit date: | 2015-06-04 | | Release date: | 2016-06-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure, mechanism, and phylogeny of LysM-chitinase conjugates specifically found in fern plants.
Plant Sci., 321, 2022
|
|
7ATK
 
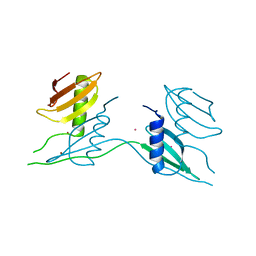 | | Crystal structure of UipA in complex with Uranium | | Descriptor: | URANIUM ATOM, UipA, ZINC ION | | Authors: | Bremond, N, Gallois, N, Legrand, P, Chapon, V, Arnoux, P. | | Deposit date: | 2020-10-30 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.855 Å) | | Cite: | Discovery and characterization of UipA, a uranium- and iron-binding PepSY protein involved in uranium tolerance by soil bacteria.
Isme J, 16, 2022
|
|
5BRO
 
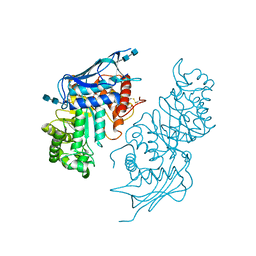 | | Crystal structure of modified HexB (modB) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-hexosaminidase subunit beta, FORMIC ACID, ... | | Authors: | Kitakaze, K, Maita, N, Itoh, K. | | Deposit date: | 2015-06-01 | | Release date: | 2016-05-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Protease-resistant modified human beta-hexosaminidase B ameliorates symptoms in GM2 gangliosidosis model.
J.Clin.Invest., 126, 2016
|
|
5B6C
 
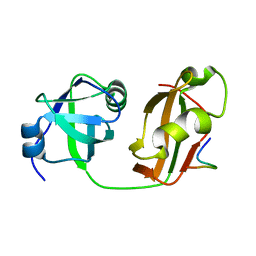 | | Structural Details of Ufd1 binding to p97 | | Descriptor: | Peptide from Ubiquitin fusion degradation protein 1 homolog, Transitional endoplasmic reticulum ATPase | | Authors: | Le, L.T.M, Yang, J.K. | | Deposit date: | 2016-05-26 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Details of Ufd1 Binding to p97 and Their Functional Implications in ER-Associated Degradation
PLoS ONE, 11, 2016
|
|
6G51
 
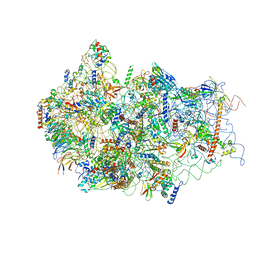 | | Cryo-EM structure of a late human pre-40S ribosomal subunit - State D | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Ameismeier, M, Cheng, J, Berninghausen, O, Beckmann, R. | | Deposit date: | 2018-03-28 | | Release date: | 2018-06-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Visualizing late states of human 40S ribosomal subunit maturation.
Nature, 558, 2018
|
|
5BCA
 
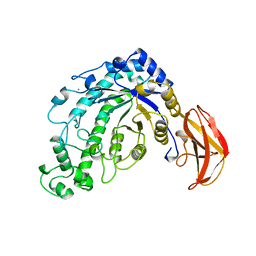 | | BETA-AMYLASE FROM BACILLUS CEREUS VAR. MYCOIDES | | Descriptor: | CALCIUM ION, PROTEIN (1,4-ALPHA-D-GLUCAN MALTOHYDROLASE.) | | Authors: | Oyama, T, Kusunoki, M, Kishimoto, Y, Takasaki, Y, Nitta, Y. | | Deposit date: | 1999-03-12 | | Release date: | 2000-03-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of beta-amylase from Bacillus cereus var. mycoides at 2.2 A resolution.
J.Biochem.(Tokyo), 125, 1999
|
|
5BKA
 
 | |
7B5A
 
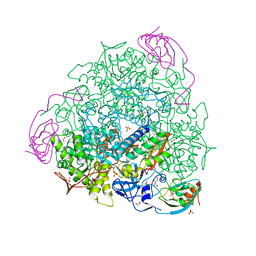 | | X-ray crystal structure of Sporosarcina pasteurii urease inhibited by Ag(PEt3)2NO3 determined at 1.97 Angstroms | | Descriptor: | 1,2-ETHANEDIOL, NICKEL (II) ION, OXYGEN ATOM, ... | | Authors: | Mazzei, L, Cianci, M, Ciurli, S. | | Deposit date: | 2020-12-03 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Kinetic and structural analysis of the inactivation of urease by mixed-ligand phosphine halide Ag(I) complexes.
J.Inorg.Biochem., 218, 2021
|
|
7B58
 
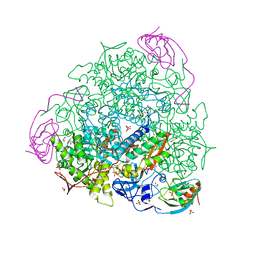 | | X-ray crystal structure of Sporosarcina pasteurii urease inhibited by Ag(PEt3)Cl determined at 1.72 Angstroms | | Descriptor: | 1,2-ETHANEDIOL, NICKEL (II) ION, OXYGEN ATOM, ... | | Authors: | Mazzei, L, Cianci, M, Ciurli, S. | | Deposit date: | 2020-12-03 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Kinetic and structural analysis of the inactivation of urease by mixed-ligand phosphine halide Ag(I) complexes.
J.Inorg.Biochem., 218, 2021
|
|
7AYX
 
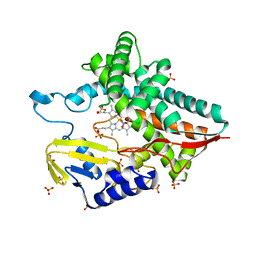 | |
7B59
 
 | | X-ray crystal structure of Sporosarcina pasteurii urease inhibited by Ag(PEt3)Br determined at 1.63 Angstroms | | Descriptor: | 1,2-ETHANEDIOL, NICKEL (II) ION, OXYGEN ATOM, ... | | Authors: | Mazzei, L, Cianci, M, Ciurli, S. | | Deposit date: | 2020-12-03 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Kinetic and structural analysis of the inactivation of urease by mixed-ligand phosphine halide Ag(I) complexes.
J.Inorg.Biochem., 218, 2021
|
|
7B0Z
 
 | |
