8RVO
 
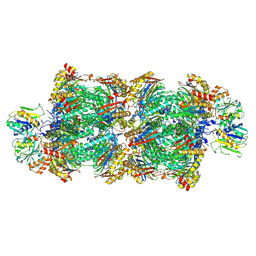 | | Proteasomal late precursor complex from pre1-1, state 1 | | Descriptor: | Probable proteasome subunit alpha type-7, Proteasome assembly chaperone 2, Proteasome chaperone 1, ... | | Authors: | Mark, E, Ramos, P.C, Kayser, F, Hoeckendorff, J, Dohmen, R.J, Wendler, P. | | Deposit date: | 2024-02-01 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Structural roles of Ump1 and beta-subunit propeptides in proteasome biogenesis.
Life Sci Alliance, 7, 2024
|
|
8VWI
 
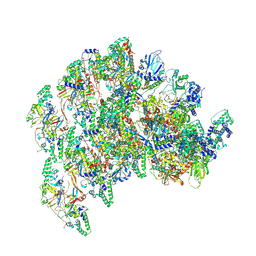 | | The base complex of the AcMNPV baculovirus nucleocapsid (Class 1, localised reconstruction) | | Descriptor: | 38K (AC98) protein in P143-LEF5 intergenic region, Capsid-associated protein VP80, Major capsid protein, ... | | Authors: | Johnstone, B.A, Koszalka, P, Ha, J, Venugopal, H, Coulibaly, F. | | Deposit date: | 2024-02-01 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.71 Å) | | Cite: | The baculovirus structure defines the hallmarks of a new viral realm
To Be Published
|
|
8RVP
 
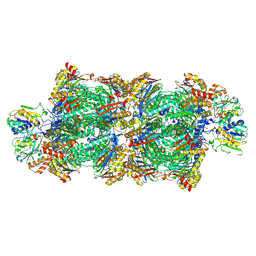 | | Proteasomal late precursor complex from pre1-1, state 2 | | Descriptor: | Probable proteasome subunit alpha type-7, Proteasome assembly chaperone 2, Proteasome chaperone 1, ... | | Authors: | Mark, E, Ramos, P.C, Kayser, F, Hoeckendorff, J, Dohmen, R.J, Wendler, P. | | Deposit date: | 2024-02-01 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.28 Å) | | Cite: | Structural roles of Ump1 and beta-subunit propeptides in proteasome biogenesis.
Life Sci Alliance, 7, 2024
|
|
8RVQ
 
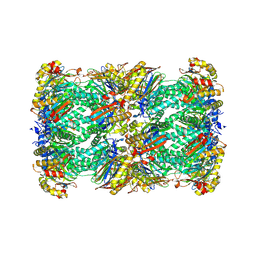 | | 20S proteasome from pre1-1 | | Descriptor: | Probable proteasome subunit alpha type-7, Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, ... | | Authors: | Mark, E, Ramos, P.C, Kayser, F, Hoeckendorff, J, Dohmen, R.J, Wendler, P. | | Deposit date: | 2024-02-01 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.02 Å) | | Cite: | Structural roles of Ump1 and beta-subunit propeptides in proteasome biogenesis.
Life Sci Alliance, 7, 2024
|
|
8RVL
 
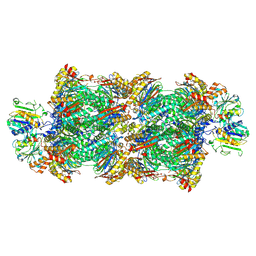 | | Proteasomal late precursor complex from pre1-1 | | Descriptor: | Probable proteasome subunit alpha type-7, Proteasome assembly chaperone 2, Proteasome chaperone 1, ... | | Authors: | Mark, E, Ramos, P.C, Kayser, F, Hoeckendorff, J, Dohmen, R.J, Wendler, P. | | Deposit date: | 2024-02-01 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.14 Å) | | Cite: | Structural roles of Ump1 and beta-subunit propeptides in proteasome biogenesis.
Life Sci Alliance, 7, 2024
|
|
8VWH
 
 | | Structure of the baculovirus major nucleocapsid protein VP39 (localised reconstruction) | | Descriptor: | Major capsid protein, ZINC ION | | Authors: | Johnstone, B.A, Hardy, J.M, Ha, J.H, Venugopal, H, Coulibaly, F. | | Deposit date: | 2024-02-01 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | The baculovirus structure defines the hallmarks of a new viral realm
To Be Published
|
|
8VWJ
 
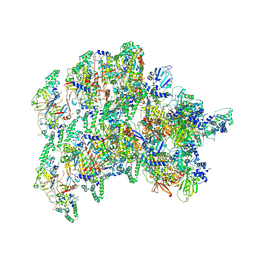 | | The base complex of the AcMNPV baculovirus nucleocapsid (Class 2, localised reconstruction) | | Descriptor: | 38K (AC98), Capsid-associated protein VP80, Major capsid protein, ... | | Authors: | Johnstone, B.A, Koszalka, P, Ha, J, Venugopal, H, Coulibaly, F. | | Deposit date: | 2024-02-01 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.78 Å) | | Cite: | The baculovirus structure defines the hallmarks of a new viral realm
To Be Published
|
|
8RVE
 
 | | Vimentin intermediate filament | | Descriptor: | Vimentin | | Authors: | Eibauer, M, Medalia, O. | | Deposit date: | 2024-02-01 | | Release date: | 2024-04-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Vimentin filaments integrate low-complexity domains in a complex helical structure.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8RUK
 
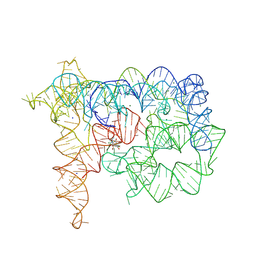 | | Structure of Oceanobacillus iheyensis group II intron in the presence of Na+, Mg2+, and ARN25850 | | Descriptor: | 2-[2,6-bis(bromanyl)-3,4,5-tris(oxidanyl)phenyl]carbonyl-~{N}-(2-pyrrolidin-1-ylethyl)-1-benzofuran-5-carboxamide, Domains 1-5, MAGNESIUM ION, ... | | Authors: | Silvestri, I, Marcia, M. | | Deposit date: | 2024-01-31 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (4.81 Å) | | Cite: | Targeting the conserved active site of splicing machines with specific and selective small molecule modulators.
Nat Commun, 15, 2024
|
|
8RUL
 
 | |
8RUN
 
 | | Structure of Oceanobacillus iheyensis group II intron in the presence of Li+, Mg2+, and ARN25850 | | Descriptor: | 2-[2,6-bis(bromanyl)-3,4,5-tris(oxidanyl)phenyl]carbonyl-~{N}-(2-pyrrolidin-1-ylethyl)-1-benzofuran-5-carboxamide, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Domains 1-5, ... | | Authors: | Silvestri, I, Marcia, M. | | Deposit date: | 2024-01-31 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (3.73 Å) | | Cite: | Targeting the conserved active site of splicing machines with specific and selective small molecule modulators.
Nat Commun, 15, 2024
|
|
8RUM
 
 | | Structure of Oceanobacillus iheyensis group II intron in the presence of Li+, Mg2+, and intronistat B | | Descriptor: | Domains 1-5, MAGNESIUM ION, ~{N}-(2-pyrrolidin-1-ylethyl)-2-[3,4,5-tris(oxidanyl)phenyl]carbonyl-1-benzofuran-5-carboxamide | | Authors: | Silvestri, I, Marcia, M. | | Deposit date: | 2024-01-31 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (3.62 Å) | | Cite: | Targeting the conserved active site of splicing machines with specific and selective small molecule modulators.
Nat Commun, 15, 2024
|
|
8RVB
 
 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 8 | | Descriptor: | (2~{R},3~{R},4~{S},5~{S})-2-(6-aminopurin-9-yl)-5-[2-(1~{H}-1,2,3-triazol-4-yl)ethylsulfanylmethyl]oxolane-3,4-diol, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kalnins, G. | | Deposit date: | 2024-01-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for inhibition of the SARS-CoV-2 nsp16 by substrate-based dual site inhibitors.
Chemmedchem, 2024
|
|
8RV8
 
 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 5 | | Descriptor: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-[[(2~{S},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]-2-chloranyl-benzoic acid, ... | | Authors: | Kalnins, G. | | Deposit date: | 2024-01-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for inhibition of the SARS-CoV-2 nsp16 by substrate-based dual site inhibitors.
Chemmedchem, 2024
|
|
8RV4
 
 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 2 | | Descriptor: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-[[(2~{S},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanylmethyl]-2-phenyl-benzoic acid, ... | | Authors: | Kalnins, G. | | Deposit date: | 2024-01-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for inhibition of the SARS-CoV-2 nsp16 by substrate-based dual site inhibitors.
Chemmedchem, 2024
|
|
8RV9
 
 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 6 | | Descriptor: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-[[(2~{S},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanylmethyl]-2-chloranyl-benzoic acid, ... | | Authors: | Kalnins, G. | | Deposit date: | 2024-01-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for inhibition of the SARS-CoV-2 nsp16 by substrate-based dual site inhibitors.
Chemmedchem, 2024
|
|
8RV5
 
 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 1 | | Descriptor: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5'-DEOXY-5'-METHYLTHIOADENOSINE, ... | | Authors: | Kalnins, G. | | Deposit date: | 2024-01-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for inhibition of the SARS-CoV-2 nsp16 by substrate-based dual site inhibitors.
Chemmedchem, 2024
|
|
8RV6
 
 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 2 | | Descriptor: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[[(2~{S},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanylmethyl]-5-(4-hydroxyphenyl)benzoic acid, ... | | Authors: | Kalnins, G. | | Deposit date: | 2024-01-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for inhibition of the SARS-CoV-2 nsp16 by substrate-based dual site inhibitors.
Chemmedchem, 2024
|
|
8RV7
 
 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 4 | | Descriptor: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[[(2~{S},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanylmethyl]-5-(3-oxidanylprop-1-ynyl)benzoic acid, ... | | Authors: | Kalnins, G. | | Deposit date: | 2024-01-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for inhibition of the SARS-CoV-2 nsp16 by substrate-based dual site inhibitors.
Chemmedchem, 2024
|
|
8RVA
 
 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 7 | | Descriptor: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[[(2~{S},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanylmethyl]benzoic acid, ... | | Authors: | Kalnins, G. | | Deposit date: | 2024-01-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for inhibition of the SARS-CoV-2 nsp16 by substrate-based dual site inhibitors.
Chemmedchem, 2024
|
|
8RUU
 
 | | Fabs derived from bimekizumab in complex with IL-17F | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoblobulin heavy chain, Immunoblobulin light chain, ... | | Authors: | Adams, R, Lawson, A.D.G. | | Deposit date: | 2024-01-31 | | Release date: | 2024-04-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Crystal Structure of Bimekizumab Fab Fragment in Complex with IL-17F Provides Molecular Basis for Dual IL-17A and IL-17F Inhibition.
J Invest Dermatol., 144, 2024
|
|
8RUH
 
 | |
8RUI
 
 | | Structure of Oceanobacillus iheyensis group II intron in the presence of K+, Mg2+, 5'-exon, and intronistat B after 1h soaking | | Descriptor: | Domains 1-5, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Silvestri, I, Marcia, M. | | Deposit date: | 2024-01-30 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Targeting the conserved active site of splicing machines with specific and selective small molecule modulators.
Nat Commun, 15, 2024
|
|
8RU8
 
 | | A crystal form of a human CDK2-CDK7 chimera | | Descriptor: | (3R,4R)-4-[[[7-[(phenylmethyl)amino]-3-propan-2-yl-pyrazolo[1,5-a]pyrimidin-5-yl]amino]methyl]piperidin-3-ol, Cyclin-dependent kinase 2 | | Authors: | Mukherjee, M, Cleasby, A. | | Deposit date: | 2024-01-30 | | Release date: | 2024-06-26 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Protein engineering enables a soakable crystal form of human CDK7 primed for high-throughput crystallography and structure-based drug design.
Structure, 32, 2024
|
|
8RUJ
 
 | | Structure of Oceanobacillus iheyensis group II intron in the presence of K+, Mg2+, 5'-exon, and ARN25850 after 1h soaking | | Descriptor: | 2-[2,6-bis(bromanyl)-3,4,5-tris(oxidanyl)phenyl]carbonyl-~{N}-(2-pyrrolidin-1-ylethyl)-1-benzofuran-5-carboxamide, Domains 1-5, MAGNESIUM ION, ... | | Authors: | Silvestri, I, Marcia, M. | | Deposit date: | 2024-01-30 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Targeting the conserved active site of splicing machines with specific and selective small molecule modulators.
Nat Commun, 15, 2024
|
|
