6FPA
 
 | | Crystal structure of anti-mTFP1 DARPin 1238_G01 in space group P212121 | | Descriptor: | DARPin 1238_G01 | | Authors: | Jakob, R.P, Vigano, M.A, Bieli, D, Matsuda, S, Schaefer, J.V, Pluckthun, A, Affolter, M, Maier, T. | | Deposit date: | 2018-02-09 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | DARPins recognizing mTFP1 as novel reagents forin vitroandin vivoprotein manipulations.
Biol Open, 7, 2018
|
|
6FP9
 
 | | Crystal structure of anti-mTFP1 DARPin 1238_E11 | | Descriptor: | 1,2-ETHANEDIOL, DARPin 1238_E11, SULFATE ION | | Authors: | Jakob, R.P, Vigano, M.A, Bieli, D, Matsuda, S, Schaefer, J.V, Pluckthun, A, Affolter, M, Maier, T. | | Deposit date: | 2018-02-09 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | DARPins recognizing mTFP1 as novel reagents forin vitroandin vivoprotein manipulations.
Biol Open, 7, 2018
|
|
6FPB
 
 | | Crystal structure of anti-mTFP1 DARPin 1238_G01 in space group I4 | | Descriptor: | CHLORIDE ION, DARPin 1238_G01 | | Authors: | Jakob, R.P, Vigano, M.A, Bieli, D, Matsuda, S, Schaefer, J.V, Pluckthun, A, Affolter, M, Maier, T. | | Deposit date: | 2018-02-09 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.617 Å) | | Cite: | DARPins recognizing mTFP1 as novel reagents forin vitroandin vivoprotein manipulations.
Biol Open, 7, 2018
|
|
6FP8
 
 | | mTFP1/DARPin 1238_E11 complex in space group C2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, DARPin1238_E11, ... | | Authors: | Jakob, R.P, Vigano, M.A, Bieli, D, Matsuda, S, Schaefer, J.V, Pluckthun, A, Affolter, M, Maier, T. | | Deposit date: | 2018-02-09 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.855 Å) | | Cite: | DARPins recognizing mTFP1 as novel reagents forin vitroandin vivoprotein manipulations.
Biol Open, 7, 2018
|
|
6BQN
 
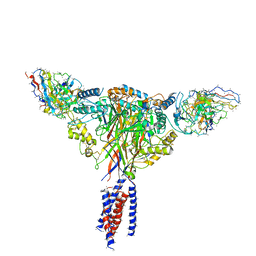 | | Cryo-EM structure of ENaC | | Descriptor: | 10D4 fab, 7B1 fab, EGFP-SCNN1G chimera, ... | | Authors: | Noreng, S, Bharadwaj, A, Posert, R, Yoshioka, C, Baconguis, I. | | Deposit date: | 2017-11-28 | | Release date: | 2018-10-10 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the human epithelial sodium channel by cryo-electron microscopy.
Elife, 7, 2018
|
|
6DQ0
 
 | | sfGFP D133 mutated to 4-nitro-L-phenylalanine | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, superfolder green fluorescent protein | | Authors: | Phillips-Piro, C.M, Maurici, N, Lee, B. | | Deposit date: | 2018-06-10 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.048 Å) | | Cite: | Crystal structures of green fluorescent protein with the unnatural amino acid 4-nitro-L-phenylalanine.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6DQ1
 
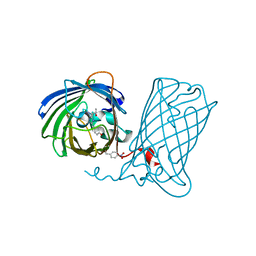 | |
6B9C
 
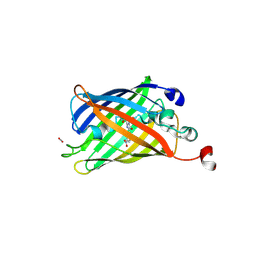 | | Superfolder Green Fluorescent Protein with 4-nitro-L-phenylalanine at the chromophore (position 66) | | Descriptor: | CARBON DIOXIDE, Green fluorescent protein | | Authors: | Phillips-Piro, C.M, Brewer, S.H, Olenginski, G.M, Piacentini, J. | | Deposit date: | 2017-10-10 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | Structural and spectrophotometric investigation of two unnatural amino-acid altered chromophores in the superfolder green fluorescent protein
Acta Crystallogr.,Sect.D, 2021
|
|
6HCQ
 
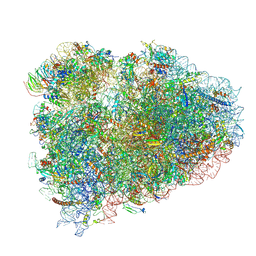 | | Structure of the rabbit collided di-ribosome (collided monosome) | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | Authors: | Juszkiewicz, S, Chandrasekaran, V, Lin, Z, Kraatz, S, Ramakrishnan, V, Hegde, R.S. | | Deposit date: | 2018-08-16 | | Release date: | 2018-10-17 | | Last modified: | 2018-11-14 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | ZNF598 Is a Quality Control Sensor of Collided Ribosomes.
Mol. Cell, 72, 2018
|
|
6HCJ
 
 | | Structure of the rabbit 80S ribosome on globin mRNA in the rotated state with A/P and P/E tRNAs | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | Authors: | Juszkiewicz, S, Chandrasekaran, V, Lin, Z, Kraatz, S, Ramakrishnan, V, Hegde, R.S. | | Deposit date: | 2018-08-15 | | Release date: | 2018-10-17 | | Last modified: | 2018-11-14 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | ZNF598 Is a Quality Control Sensor of Collided Ribosomes.
Mol. Cell, 72, 2018
|
|
6HCM
 
 | | Structure of the rabbit collided di-ribosome (stalled monosome) | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | Authors: | Juszkiewicz, S, Chandrasekaran, V, Lin, Z, Kraatz, S, Ramakrishnan, V, Hegde, R.S. | | Deposit date: | 2018-08-15 | | Release date: | 2018-10-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | ZNF598 Is a Quality Control Sensor of Collided Ribosomes.
Mol. Cell, 72, 2018
|
|
6HCF
 
 | | Structure of the rabbit 80S ribosome stalled on globin mRNA at the stop codon | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | Authors: | Juszkiewicz, S, Chandrasekaran, V, Lin, Z, Kraatz, S, Ramakrishnan, V, Hegde, R.S. | | Deposit date: | 2018-08-14 | | Release date: | 2018-10-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | ZNF598 Is a Quality Control Sensor of Collided Ribosomes.
Mol. Cell, 72, 2018
|
|
6HWP
 
 | |
6MKS
 
 | | Cryo-EM structure of NLRC4-CARD filament | | Descriptor: | Chimera protein of NLR family CARD domain-containing protein 4 and EGFP | | Authors: | Zheng, W, Matyszewski, M, Sohn, J, Egelman, E.H. | | Deposit date: | 2018-09-26 | | Release date: | 2018-11-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the NLRC4CARDfilament provides insights into how symmetric and asymmetric supramolecular structures drive inflammasome assembly.
J. Biol. Chem., 293, 2018
|
|
6MTB
 
 | | Rabbit 80S ribosome with P- and Z-site tRNAs (unrotated state) | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Brown, A, Baird, M.R, Yip, M.C.J, Murray, J, Shao, S. | | Deposit date: | 2018-10-19 | | Release date: | 2018-11-21 | | Last modified: | 2019-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of translationally inactive mammalian ribosomes.
Elife, 7, 2018
|
|
6MTD
 
 | | Rabbit 80S ribosome with eEF2 and SERBP1 (unrotated state with 40S head swivel) | | Descriptor: | 18S rRNA, 28S rRNA, 5.8S rRNA, ... | | Authors: | Brown, A, Baird, M.R, Yip, M.C.J, Murray, J, Shao, S. | | Deposit date: | 2018-10-19 | | Release date: | 2018-11-21 | | Last modified: | 2019-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of translationally inactive mammalian ribosomes.
Elife, 7, 2018
|
|
6MWQ
 
 | |
6MTE
 
 | | Rabbit 80S ribosome with eEF2 and SERBP1 (rotated state) | | Descriptor: | 18S rRNA, 28S rRNA, 5.8S rRNA, ... | | Authors: | Brown, A, Baird, M.R, Yip, M.C.J, Murray, J, Shao, S. | | Deposit date: | 2018-10-19 | | Release date: | 2018-11-21 | | Last modified: | 2019-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of translationally inactive mammalian ribosomes.
Elife, 7, 2018
|
|
6MTC
 
 | | Rabbit 80S ribosome with Z-site tRNA and IFRD2 (unrotated state) | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Brown, A, Baird, M.R, Yip, M.C.J, Murray, J, Shao, S. | | Deposit date: | 2018-10-19 | | Release date: | 2018-11-21 | | Last modified: | 2019-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of translationally inactive mammalian ribosomes.
Elife, 7, 2018
|
|
6GZ3
 
 | | tRNA translocation by the eukaryotic 80S ribosome and the impact of GTP hydrolysis, Translocation-intermediate-POST-1 (TI-POST-1) | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | Authors: | Flis, J, Holm, M, Rundlet, E.J, Loerke, J, Hilal, T, Dabrowski, M, Buerger, J, Mielke, T, Blanchard, S.C, Spahn, C.M.T, Budkevich, T.V. | | Deposit date: | 2018-07-03 | | Release date: | 2018-12-05 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | tRNA Translocation by the Eukaryotic 80S Ribosome and the Impact of GTP Hydrolysis.
Cell Rep, 25, 2018
|
|
6GZ4
 
 | | tRNA translocation by the eukaryotic 80S ribosome and the impact of GTP hydrolysis, Translocation-intermediate-POST-2 (TI-POST-2) | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Flis, J, Holm, M, Rundlet, E.J, Loerke, J, Hilal, T, Dabrowski, M, Buerger, J, Mielke, T, Blanchard, S.C, Spahn, C.M.T, Budkevich, T.V. | | Deposit date: | 2018-07-03 | | Release date: | 2018-12-05 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | tRNA Translocation by the Eukaryotic 80S Ribosome and the Impact of GTP Hydrolysis.
Cell Rep, 25, 2018
|
|
6FYX
 
 | | Structure of a partial yeast 48S preinitiation complex with eIF5 N-terminal domain (model C1) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0, 40S ribosomal protein S1, ... | | Authors: | Llacer, J.L, Hussain, T, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2018-03-12 | | Release date: | 2018-12-05 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Translational initiation factor eIF5 replaces eIF1 on the 40S ribosomal subunit to promote start-codon recognition.
Elife, 7, 2018
|
|
6FYY
 
 | | Structure of a partial yeast 48S preinitiation complex with eIF5 N-terminal domain (model C2) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0, 40S ribosomal protein S1, ... | | Authors: | Llacer, J.L, Hussain, T, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2018-03-12 | | Release date: | 2018-12-05 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Translational initiation factor eIF5 replaces eIF1 on the 40S ribosomal subunit to promote start-codon recognition.
Elife, 7, 2018
|
|
6GZ5
 
 | | tRNA translocation by the eukaryotic 80S ribosome and the impact of GTP hydrolysis, Translocation-intermediate-POST-3 (TI-POST-3) | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Flis, J, Holm, M, Rundlet, E.J, Loerke, J, Hilal, T, Dabrowski, M, Buerger, J, Mielke, T, Blanchard, S.C, Spahn, C.M.T, Budkevich, T.V. | | Deposit date: | 2018-07-03 | | Release date: | 2018-12-05 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | tRNA Translocation by the Eukaryotic 80S Ribosome and the Impact of GTP Hydrolysis.
Cell Rep, 25, 2018
|
|
6F5J
 
 | |
