6RHC
 
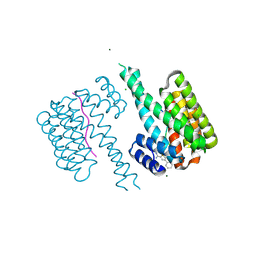 | | Fragment AZ-003 binding at the TAZpS89/14-3-3 sigma interface | | Descriptor: | 14-3-3 protein sigma, 5-azanyl-4-phenyl-thiophene-2-carboximidamide, CHLORIDE ION, ... | | Authors: | Genet, S, Wolter, M, Guillory, X, Somsen, B, Leysen, S, Castaldi, P, Ottmann, C. | | Deposit date: | 2019-04-19 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
6RSR
 
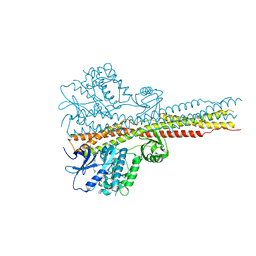 | | TBK1 in complex with compound 2 | | Descriptor: | Serine/threonine-protein kinase TBK1, ~{N}-(cyclopropen-1-ylmethyl)-2-[[4-[[4-[3,3,3-tris(fluoranyl)propanoyl]piperazin-1-yl]methyl]pyridin-2-yl]amino]-1~{H}-benzimidazole-5-carboxamide | | Authors: | Panne, D, Hillig, R.C, Rengachari, S. | | Deposit date: | 2019-05-22 | | Release date: | 2020-01-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Discovery of BAY-985, a Highly Selective TBK1/IKK epsilon Inhibitor.
J.Med.Chem., 63, 2020
|
|
2B01
 
 | | Crystal Structure of Porcine Pancreatic Phospholipase A2 in Complex with Taurochenodeoxycholate | | Descriptor: | CALCIUM ION, CHLORIDE ION, Phospholipase A2, ... | | Authors: | Pan, Y.H, Bahnson, B.J, Jain, M.K. | | Deposit date: | 2005-09-12 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for bile salt inhibition of pancreatic phospholipase A2.
J.Mol.Biol., 369, 2007
|
|
2B00
 
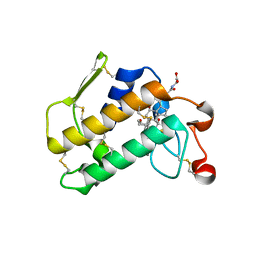 | | Crystal Structure of Porcine Pancreatic Phospholipase A2 in Complex with Glycocholate | | Descriptor: | CALCIUM ION, GLYCOCHOLIC ACID, Phospholipase A2, ... | | Authors: | Pan, Y.H, Bahnson, B.J, Jain, M.K. | | Deposit date: | 2005-09-12 | | Release date: | 2006-11-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for bile salt inhibition of pancreatic phospholipase A2.
J.Mol.Biol., 369, 2007
|
|
2B04
 
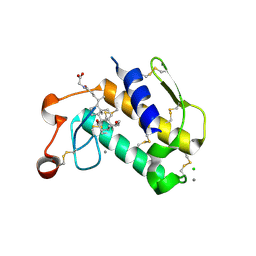 | | Crystal Structure of Porcine Pancreatic Phospholipase A2 in Complex with Glycochenodeoxycholate | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCOCHENODEOXYCHOLIC ACID, ... | | Authors: | Pan, Y.H, Bahnson, B.J, Jain, M.K. | | Deposit date: | 2005-09-12 | | Release date: | 2006-11-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for bile salt inhibition of pancreatic phospholipase A2.
J.Mol.Biol., 369, 2007
|
|
6TA2
 
 | | Human NAMPT in complex with nicotinic acid mononucleotide and phosphate | | Descriptor: | CHLORIDE ION, GLYCEROL, NICOTINATE MONONUCLEOTIDE, ... | | Authors: | Houry, D, Raasakka, A, Kursula, P, Ziegler, M. | | Deposit date: | 2019-10-29 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Identification of structural determinants of NAMPT activity and substrate selectivity
To Be Published
|
|
6T5F
 
 | | Human 14-3-3 sigma fused to the StARD1 peptide including phosphoserine-195 | | Descriptor: | 14-3-3 protein sigma, StARD1 peptide | | Authors: | Sluchanko, N.N, Tugaeva, K.V, Titterington, J, Antson, A.A. | | Deposit date: | 2019-10-16 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Molecular basis for the recognition of steroidogenic acute regulatory protein by the 14-3-3 protein family.
Febs J., 287, 2020
|
|
1TJZ
 
 | |
6TAC
 
 | | Human NAMPT deletion mutant in complex with nicotinamide mononucleotide, pyrophosphate, and Mg2+ | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Houry, D, Raasakka, A, Kursula, P, Ziegler, M. | | Deposit date: | 2019-10-29 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of structural determinants of NAMPT activity and substrate selectivity
To Be Published
|
|
6T80
 
 | |
2AZY
 
 | | Crystal Structure of Porcine Pancreatic Phospholipase A2 in Complex with Cholate | | Descriptor: | CALCIUM ION, CHOLIC ACID, Phospholipase A2, ... | | Authors: | Pan, Y.H, Bahnson, B.J, Jain, M.K. | | Deposit date: | 2005-09-12 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for bile salt inhibition of pancreatic phospholipase A2.
J.Mol.Biol., 369, 2007
|
|
5VF9
 
 | | Native human manganese superoxide dismutase | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Azadmanesh, J, Trickel, S.R, Borgstahl, G.E.O. | | Deposit date: | 2017-04-07 | | Release date: | 2017-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Substrate-analog binding and electrostatic surfaces of human manganese superoxide dismutase.
J. Struct. Biol., 199, 2017
|
|
1BUY
 
 | | HUMAN ERYTHROPOIETIN, NMR MINIMIZED AVERAGE STRUCTURE | | Descriptor: | PROTEIN (ERYTHROPOIETIN) | | Authors: | Cheetham, J.C, Smith, D.M, Aoki, K.H, Stevenson, J.L, Hoeffel, T.J, Syed, R.S, Egrie, J, Harvey, T.S. | | Deposit date: | 1998-09-08 | | Release date: | 1999-09-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of human erythropoietin and a comparison with its receptor bound conformation.
Nat.Struct.Biol., 5, 1998
|
|
5T30
 
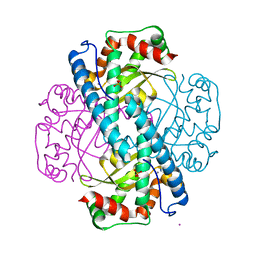 | | Human MnSOD-azide complex | | Descriptor: | AZIDE ION, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Azadmanesh, J, Trickel, S.R, Kolar, C.H, Lovelace, J.J, Borgstahl, G.E.O. | | Deposit date: | 2016-08-24 | | Release date: | 2017-05-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Substrate-analog binding and electrostatic surfaces of human manganese superoxide dismutase.
J. Struct. Biol., 199, 2017
|
|
2O25
 
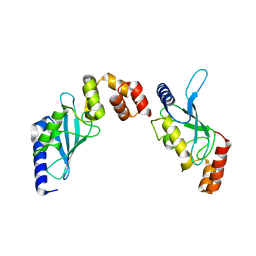 | | Ubiquitin-Conjugating Enzyme E2-25 kDa Complexed With SUMO-1-Conjugating Enzyme UBC9 | | Descriptor: | SUMO-1-conjugating enzyme UBC9, Ubiquitin-conjugating enzyme E2-25 kDa | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Newman, E.M, Mackenzie, F, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-11-29 | | Release date: | 2007-01-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Novel and Unexpected Complex Between the SUMO-1-Conjugating Enzyme UBC9 and the Ubiquitin-Conjugating Enzyme E2-25 kDa
To be Published
|
|
3EWF
 
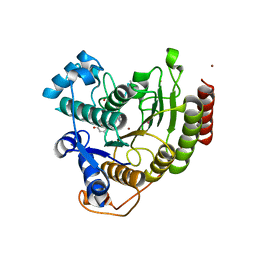 | | Crystal Structure Analysis of human HDAC8 H143A variant complexed with substrate. | | Descriptor: | 7-AMINO-4-METHYL-CHROMEN-2-ONE, Histone deacetylase 8, PEPTIDIC SUBSTRATE, ... | | Authors: | Dowling, D.P, Gantt, S.L, Gattis, S.G, Fierke, C.A, Christianson, D.W. | | Deposit date: | 2008-10-14 | | Release date: | 2008-12-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural studies of human histone deacetylase 8 and its site-specific variants complexed with substrate and inhibitors.
Biochemistry, 47, 2008
|
|
3SD3
 
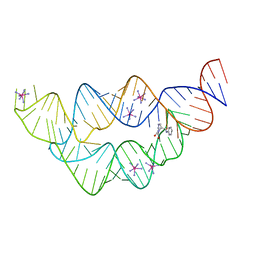 | | The structure of the tetrahydrofolate riboswitch containing a U25C mutation | | Descriptor: | IRIDIUM HEXAMMINE ION, N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, Tetrahydrofolate riboswitch | | Authors: | Reyes, F.E, Trausch, J.J, Ceres, P, Batey, R.T. | | Deposit date: | 2011-06-08 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structure of a tetrahydrofolate-sensing riboswitch reveals two ligand binding sites in a single aptamer.
Structure, 19, 2011
|
|
4G5I
 
 | | Crystal Structure of Porcine pancreatic PlA2 in complex with DBP | | Descriptor: | CALCIUM ION, CHLORIDE ION, Phospholipase A2, ... | | Authors: | Prasanth, G.K, Naveen, C.D, Mandal, P.K, Karthe, P, Haridas, M, Sadasivan, C. | | Deposit date: | 2012-07-18 | | Release date: | 2012-11-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Porcine pancreatic PlA2 in complex with DBP
TO BE PUBLISHED
|
|
7CWE
 
 | | Human Fructose-1,6-bisphosphatase 1 in APO R-state | | Descriptor: | Fructose-1,6-bisphosphatase 1, MAGNESIUM ION | | Authors: | Chen, Y, Zhang, J, Li, C, Cao, Y. | | Deposit date: | 2020-08-28 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Human Fructose-1,6-bisphosphatase 1 in APO R-state
To Be Published
|
|
7CVH
 
 | | Human Fructose-1,6-bisphosphatase 1 in complex with geranylgeranyl diphosphate | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, Fructose-1,6-bisphosphatase 1, ... | | Authors: | Chen, Y, Zhang, J, Li, C, Cao, Y. | | Deposit date: | 2020-08-26 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The structural basis for GGPP activation on the enzymatic activities FBP1
To Be Published
|
|
1FPT
 
 | |
1B44
 
 | | CRYSTAL STRUCTURE OF THE B SUBUNIT OF HEAT-LABILE ENTEROTOXIN FROM E. COLI CARRYING A PEPTIDE WITH ANTI-HSV ACTIVITY | | Descriptor: | PROTEIN (B-POL SUBUNIT OF HEAT-LABILE ENTEROTOXIN) | | Authors: | Matkovic-Calogovic, D, Loregian, A, D'Acunto, M.R, Battistutta, R, Tossi, A, Palu, G, Zanotti, G. | | Deposit date: | 1999-01-04 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of the B subunit of Escherichia coli heat-labile enterotoxin carrying peptides with anti-herpes simplex virus type 1 activity.
J.Biol.Chem., 274, 1999
|
|
1AWI
 
 | |
8SWZ
 
 | | PARP4 ART domain bound to EB47 | | Descriptor: | 2-[4-[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]carbonylpiperazin-1-yl]-N-(1-oxidanylidene-2,3-dihydroisoindol-4-yl)ethanamide, GLYCEROL, Protein mono-ADP-ribosyltransferase PARP4 | | Authors: | Frigon, L, Pascal, J.M. | | Deposit date: | 2023-05-19 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and biochemical analysis of the PARP1-homology region of PARP4/vault PARP.
Nucleic Acids Res., 51, 2023
|
|
8SX2
 
 | | PARP4 catalytic domain bound to EB47 | | Descriptor: | 2-[4-[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]carbonylpiperazin-1-yl]-N-(1-oxidanylidene-2,3-dihydroisoindol-4-yl)ethanamide, Protein mono-ADP-ribosyltransferase PARP4 | | Authors: | Frigon, L, Pascal, J.M. | | Deposit date: | 2023-05-19 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural and biochemical analysis of the PARP1-homology region of PARP4/vault PARP.
Nucleic Acids Res., 51, 2023
|
|
