6CTB
 
 | |
2M36
 
 | |
1F03
 
 | | SOLUTION STRUCTURE OF OXIDIZED BOVINE MICROSOMAL CYTOCHROME B5 MUTANT (E44A, E48A, E56A, D60A) AND ITS INTERACTION WITH CYTOCHROME C | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wu, Y.B, Lu, J, Qian, C.M, Tang, W.X, Li, E.C, Wang, J.F, Wang, Y.H, Wang, W.H, Lu, J.X, Xie, Y, Huang, Z.X. | | Deposit date: | 2000-05-14 | | Release date: | 2000-06-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of cytochrome b(5) mutant (E44/48/56A/D60A) and its interaction with cytochrome c.
Eur.J.Biochem., 268, 2001
|
|
1F04
 
 | | SOLUTION STRUCTURE OF OXIDIZED BOVINE MICROSOMAL CYTOCHROME B5 MUTANT (E44A, E48A, E56A, D60A) AND ITS INTERACTION WITH CYTOCHROME C | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wu, Y.B, Lu, J, Qian, C.M, Tang, W.X, Li, E.C, Wang, J.F, Wang, Y.H, Wang, W.H, Lu, J.X, Xie, Y, Huang, Z.X. | | Deposit date: | 2000-05-14 | | Release date: | 2000-06-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of cytochrome b(5) mutant (E44/48/56A/D60A) and its interaction with cytochrome c.
Eur.J.Biochem., 268, 2001
|
|
1FKC
 
 | | HUMAN PRION PROTEIN (MUTANT E200K) FRAGMENT 90-231 | | Descriptor: | PRION PROTEIN | | Authors: | Zhang, Y, Swietnicki, W, Zagorski, M.G, Surewicz, W.K, Soennichsen, F.D. | | Deposit date: | 2000-08-09 | | Release date: | 2000-09-21 | | Last modified: | 2018-03-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the E200K variant of human prion protein. Implications for the mechanism of pathogenesis in familial prion diseases.
J.Biol.Chem., 275, 2000
|
|
1FSH
 
 | |
1GKG
 
 | | Structure Determination and Rational Mutagenesis reveal binding surface of immune adherence receptor, CR1 (CD35) | | Descriptor: | COMPLEMENT RECEPTOR TYPE 1 | | Authors: | Smith, B.O, Mallin, R.L, Krych-Goldberg, M, Wang, X, Hauhart, R.E, Bromek, K, Uhrin, D, Atkinson, J.P, Barlow, P.N. | | Deposit date: | 2001-08-14 | | Release date: | 2002-04-18 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure of the C3B Binding Site of Cr1 (Cd35), the Immune Adherence Receptor
Cell(Cambridge,Mass.), 108, 2002
|
|
1GD5
 
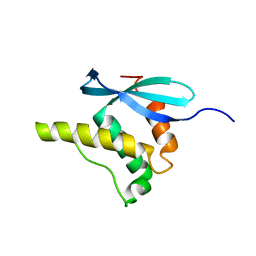 | | SOLUTION STRUCTURE OF THE PX DOMAIN FROM HUMAN P47PHOX NADPH OXIDASE | | Descriptor: | NEUTROPHIL CYTOSOL FACTOR 1 | | Authors: | Hiroaki, H, Ago, T, Ito, T, Sumimoto, H, Kohda, D. | | Deposit date: | 2000-09-14 | | Release date: | 2001-06-13 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PX domain, a target of the SH3 domain.
Nat.Struct.Biol., 8, 2001
|
|
1GKN
 
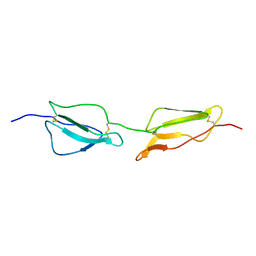 | | Structure Determination and Rational Mutagenesis reveal binding surface of immune adherence receptor, CR1 (CD35) | | Descriptor: | COMPLEMENT RECEPTOR TYPE 1 | | Authors: | Smith, B.O, Mallin, R.L, Krych-Goldberg, M, Wang, X, Hauhart, R.E, Bromek, K, Uhrin, D, Atkinson, J.P, Barlow, P.N. | | Deposit date: | 2001-08-16 | | Release date: | 2002-04-18 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure of the C3B Binding Site of Cr1 (Cd35), the Immune Adherence Receptor
Cell(Cambridge,Mass.), 108, 2002
|
|
1GM1
 
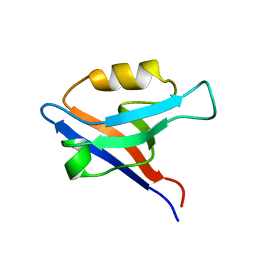 | | Second PDZ Domain (PDZ2) of PTP-BL | | Descriptor: | PROTEIN TYROSINE PHOSPHATASE | | Authors: | Walma, T, Tessari, M, Aelen, J, Schepens, J, Hendriks, W, Vuister, G.W. | | Deposit date: | 2001-09-06 | | Release date: | 2002-03-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure, dynamics and binding characteristics of the second PDZ domain of PTP-BL.
J. Mol. Biol., 316, 2002
|
|
1H0J
 
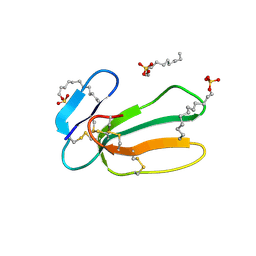 | | Structural Basis of the Membrane-induced Cardiotoxin A3 Oligomerization | | Descriptor: | CARDIOTOXIN-3, DODECYL SULFATE | | Authors: | Forouhar, F, Huang, W.-N, Liu, J.-H, Chien, K.-Y, Wu, W.-G, Hsiao, C.-D. | | Deposit date: | 2002-06-20 | | Release date: | 2003-06-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of Membrane-Induced Cardiotoxin A3 Oligomerization
J.Biol.Chem., 278, 2003
|
|
1GO5
 
 | |
1H3H
 
 | | Structural Basis for Specific Recognition of an RxxK-containing SLP-76 peptide by the Gads C-terminal SH3 domain | | Descriptor: | GRB2-RELATED ADAPTOR PROTEIN 2, LYMPHOCYTE CYTOSOLIC PROTEIN 2 | | Authors: | Liu, Q, Berry, D, Nash, P, Pawson, T, McGlade, C.J, Li, S.S. | | Deposit date: | 2002-09-03 | | Release date: | 2003-03-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Specific Binding of the Gads SH3 Domain to an Rxxk Motif-Containing Slp-76 Peptide: A Novel Mode of Peptide Recognition
Mol.Cell, 11, 2003
|
|
1H7Y
 
 | | Translationally Controlled Tumor-associated Protein p23fyp from Schizosaccharomyces pombe | | Descriptor: | TRANSLATIONALLY CONTROLLED TUMOR PROTEIN | | Authors: | Thaw, P, Baxter, N.J, Sedelnikova, S.E, Price, C, Waltho, J.P, Craven, C.J. | | Deposit date: | 2001-01-19 | | Release date: | 2001-08-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of TCTP reveals unexpected relationship with guanine nucleotide-free chaperones.
Nat. Struct. Biol., 8, 2001
|
|
1H6Q
 
 | | Translationally Controlled Tumor-associated Protein p23fyp from Schizosaccharomyces pombe | | Descriptor: | TRANSLATIONALLY CONTROLLED TUMOR PROTEIN | | Authors: | Thaw, P, Baxter, N.J, Sedelnikova, S.E, Price, C, Waltho, J.P, Craven, C.J. | | Deposit date: | 2001-06-20 | | Release date: | 2001-08-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of TCTP reveals unexpected relationship with guanine nucleotide-free chaperones.
Nat. Struct. Biol., 8, 2001
|
|
1D4B
 
 | | CIDE-N DOMAIN OF HUMAN CIDE-B | | Descriptor: | HUMAN CELL DEATH-INDUCING EFFECTOR B | | Authors: | Lugovskoy, A, Zhou, P, Chou, J, McCarty, J, Li, P, Wagner, G. | | Deposit date: | 1999-10-02 | | Release date: | 1999-12-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the CIDE-N domain of CIDE-B and a model for CIDE-N/CIDE-N interactions in the DNA fragmentation pathway of apoptosis.
Cell(Cambridge,Mass.), 99, 1999
|
|
1HG7
 
 | | High resolution structure of HPLC-12 type III antifreeze protein from Ocean Pout Macrozoarces americanus | | Descriptor: | HPLC-12 TYPE III ANTIFREEZE PROTEIN, SULFATE ION | | Authors: | Antson, A.A, Smith, D.J, Roper, D.I, Lewis, S, Caves, L.S.D, Verma, C.S, Buckley, S.L, Lillford, P.J, Hubbard, R.E. | | Deposit date: | 2000-12-13 | | Release date: | 2001-01-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Understanding the Mechanism of Ice Binding by Type III Antifreeze Protein
J.Mol.Biol., 305, 2001
|
|
1H1X
 
 | | Sperm whale Myoglobin mutant T67R S92D | | Descriptor: | CYANIDE ION, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Zuccotti, S, Bolognesi, M. | | Deposit date: | 2002-07-25 | | Release date: | 2003-10-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Engineering Peroxidase Activity in Myoglobin: The Haem Cavity Structure and Peroxide Activation in the T67R/S92D Mutant and its Derivative Reconstituted with Protohaemin-L-Histidine.
Biochem.J., 377, 2004
|
|
1HAQ
 
 | |
1HHV
 
 | | SOLUTION STRUCTURE OF VIRUS CHEMOKINE VMIP-II | | Descriptor: | VIRUS CHEMOKINE VMIP-II | | Authors: | Shao, W, Fernandez, E, Navenot, J.M, Wilken, J, Thompson, D.A, Pepiper, S, Schweitzer, B.I, Lolis, E. | | Deposit date: | 1998-12-06 | | Release date: | 2003-09-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | CCR2 and CCR5 receptor-binding properties of herpesvirus-8
vMIP-II based on sequence analysis and its solution structure
Eur.J.Biochem., 268, 2001
|
|
1HRE
 
 | | SOLUTION STRUCTURE OF THE EPIDERMAL GROWTH FACTOR-LIKE DOMAIN OF HEREGULIN-ALPHA, A LIGAND FOR P180ERB4 | | Descriptor: | HEREGULIN ALPHA | | Authors: | Nagata, K, Kohda, D, Hatanaka, H, Ichikawa, S, Inagaki, F. | | Deposit date: | 1994-07-21 | | Release date: | 1994-10-15 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the epidermal growth factor-like domain of heregulin-alpha, a ligand for p180erbB-4.
EMBO J., 13, 1994
|
|
1HRF
 
 | | SOLUTION STRUCTURE OF THE EPIDERMAL GROWTH FACTOR-LIKE DOMAIN OF HEREGULIN-ALPHA, A LIGAND FOR P180ERB4 | | Descriptor: | HEREGULIN ALPHA | | Authors: | Nagata, K, Kohda, D, Hatanaka, H, Ichikawa, S, Inagaki, F. | | Deposit date: | 1994-07-21 | | Release date: | 1994-10-15 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the epidermal growth factor-like domain of heregulin-alpha, a ligand for p180erbB-4.
EMBO J., 13, 1994
|
|
1C3T
 
 | |
1DZ1
 
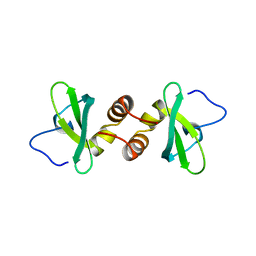 | | Mouse HP1 (M31) C terminal (shadow chromo) domain | | Descriptor: | MODIFIER 1 PROTEIN | | Authors: | Brasher, S.V, Smith, B.O, Fogh, R.H, Nietlispach, D, Thiru, A, Nielsen, P.R, Broadhurst, R.W, Ball, L.J, Murzina, N, Laue, E.D. | | Deposit date: | 2000-02-11 | | Release date: | 2000-04-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Structure of Mouse Hp1 Suggests a Unique Mode of Single Peptide Recognition by the Shadow Chromo Domain Dimer
Embo J., 19, 2000
|
|
3ZEH
 
 | |
