4JON
 
 | |
4GU6
 
 | | FOCAL ADHESION KINASE CATALYTIC DOMAIN IN COMPLEX WITH N-{3-[(5-Cyano-2-phenyl-1H-pyrrolo[2,3-b]pyridin-4-ylamino)- methyl]-pyridin-2-yl}-N-methyl-methanesulfonamide | | Descriptor: | Focal adhesion kinase 1, N-(3-{[(5-cyano-2-phenyl-1H-pyrrolo[2,3-b]pyridin-4-yl)amino]methyl}pyridin-2-yl)-N-methylmethanesulfonamide | | Authors: | Musil, D, Heinrich, T. | | Deposit date: | 2012-08-29 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Fragment-based discovery of new highly substituted 1H-pyrrolo[2,3-b]- and 3H-imidazolo[4,5-b]-pyridines as focal adhesion kinase inhibitors.
J.Med.Chem., 56, 2013
|
|
4HFQ
 
 | | Crystal structure of UDP-X diphosphatase | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Duong-Ly, K.C, Amzel, L.M, Gabelli, S.B. | | Deposit date: | 2012-10-05 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | A UDP-X diphosphatase from Streptococcus pneumoniae hydrolyzes precursors of peptidoglycan biosynthesis.
Plos One, 8, 2013
|
|
4HBR
 
 | |
4HF7
 
 | |
4GOT
 
 | |
4H3P
 
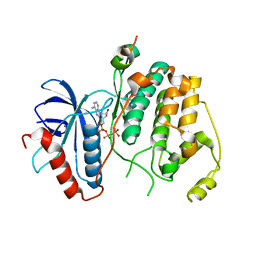 | | Crystal structure of human ERK2 complexed with a MAPK docking peptide | | Descriptor: | Mitogen-activated protein kinase 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Ribosomal protein S6 kinase alpha-1 | | Authors: | Gogl, G, Toeroe, I, Remenyi, A. | | Deposit date: | 2012-09-14 | | Release date: | 2013-02-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Protein-peptide complex crystallization: a case study on the ERK2 mitogen-activated protein kinase
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4JX0
 
 | |
4JH0
 
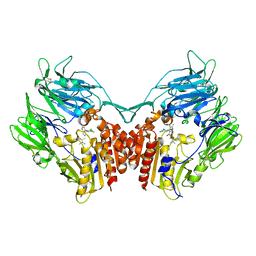 | | Crystal structure of dipeptidyl-peptidase 4 (CD26, adenosine deaminase complexing protein 2) (DPP-IV-WT) complex with bms-767778 AKA 2-(3-(aminomethyl)-4-(2,4- dichlorophenyl)-2-methyl-5-oxo-5,7-dihydro-6h-pyrrolo[3,4- b]pyridin-6-yl)-n,n-dimethylacetamide | | Descriptor: | 2-[3-(aminomethyl)-4-(2,4-dichlorophenyl)-2-methyl-5-oxo-5,7-dihydro-6H-pyrrolo[3,4-b]pyridin-6-yl]-N,N-dimethylacetamide, Dipeptidyl peptidase 4 | | Authors: | Klei, H.E. | | Deposit date: | 2013-03-04 | | Release date: | 2013-09-04 | | Last modified: | 2013-10-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Optimization of Activity, Selectivity, and Liability Profiles in 5-Oxopyrrolopyridine DPP4 Inhibitors Leading to Clinical Candidate (Sa)-2-(3-(Aminomethyl)-4-(2,4-dichlorophenyl)-2-methyl-5-oxo-5H-pyrrolo[3,4-b]pyridin-6(7H)-yl)-N,N-dimethylacetamide (BMS-767778).
J.Med.Chem., 56, 2013
|
|
4JNM
 
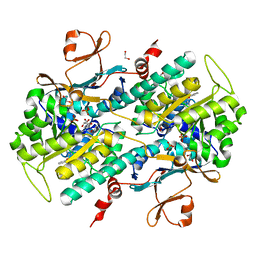 | | Discovery of Potent and Efficacious Urea-containing Nicotinamide Phosphoribosyltransferase (NAMPT) Inhibitors with Reduced CYP2C9 Inhibition Properties | | Descriptor: | 1,2-ETHANEDIOL, 1-[(6-aminopyridin-3-yl)methyl]-3-[4-(phenylsulfonyl)phenyl]urea, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Gunzner-Toste, J, Zhao, G, Bauer, P, Baumeister, T, Buckmelter, A.J, Caligiuri, M, Clodfelter, K.H, Fu, B, Han, B, Ho, Y, Kley, N, Liederer, B, Lin, J, Mukadam, S, O'Brien, T, Reynolds, D.J, Sharma, G, Skelton, N, Smith, C.C, Oh, A, Wang, W, Wang, Z, Xiao, Y, Yuen, P, Zak, M, Zhang, L, Zheng, X, Bair, K.W, Dragovich, P.S. | | Deposit date: | 2013-03-15 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of potent and efficacious urea-containing nicotinamide phosphoribosyltransferase (NAMPT) inhibitors with reduced CYP2C9 inhibition properties.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4JQ0
 
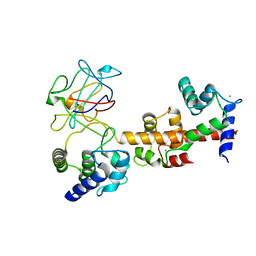 | | Voltage-gated sodium channel 1.5 C-terminal domain in complex with FGF12B and Ca2+/calmodulin | | Descriptor: | CALCIUM ION, Calmodulin, Fibroblast growth factor 12, ... | | Authors: | Wang, C, Chung, B.C, Yan, H, Wang, H.G, Lee, S.Y, Pitt, G.S. | | Deposit date: | 2013-03-19 | | Release date: | 2014-05-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.84 Å) | | Cite: | Structural analyses of Ca(2+)/CaM interaction with NaV channel C-termini reveal mechanisms of calcium-dependent regulation.
Nat Commun, 5, 2014
|
|
4JX2
 
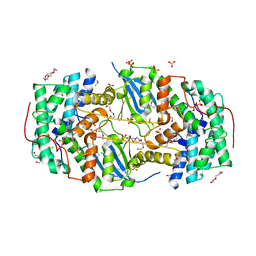 | |
4KAO
 
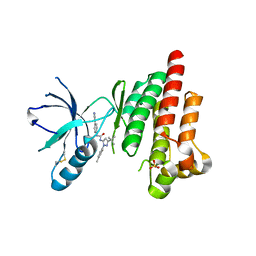 | | FOCAL ADHESION KINASE CATALYTIC DOMAIN IN COMPLEX WITH 1-(5-tert-Butyl-2-p-tolyl-2H-pyrazol-3-yl)-3-(4-pyridin-3- yl-phenyl)-urea | | Descriptor: | 1-[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]-3-[4-(pyridin-3-yl)phenyl]urea, Focal adhesion kinase 1, SULFATE ION | | Authors: | Musil, D, Graedler, U, Heinrich, T, Lehmann, M, Dresing, V. | | Deposit date: | 2013-04-22 | | Release date: | 2013-09-11 | | Last modified: | 2013-10-09 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Fragment-based discovery of focal adhesion kinase inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4KIE
 
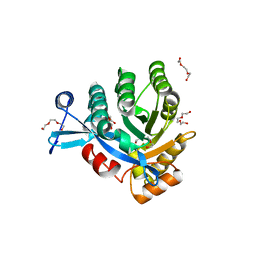 | |
4KNI
 
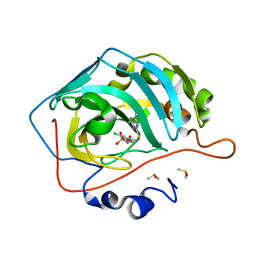 | | Crystal structure of human carbonic anhydrase isozyme II with 2-Chloro-4-{[(4,6-dimethylpyrimidin-2-yl)sulfanyl]acetyl}benzenesulfonamide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-chloro-4-{[(4,6-dimethylpyrimidin-2-yl)sulfanyl]acetyl}benzenesulfonamide, Carbonic anhydrase 2, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2013-05-10 | | Release date: | 2013-11-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Benzenesulfonamides with pyrimidine moiety as inhibitors of human carbonic anhydrases I, II, VI, VII, XII, and XIII
Bioorg.Med.Chem., 21, 2013
|
|
4K0O
 
 | | F17b-G lectin domain with bound GlcNAc(beta1-3)Gal | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-methyl beta-D-galactopyranoside, F17b-G fimbrial adhesin, NICKEL (II) ION, ... | | Authors: | Buts, L, Loris, R, Bouckaert, J, Moonens, K. | | Deposit date: | 2013-04-04 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Sampling of Glycan Interaction Profiles Reveals Mucosal Receptors for Fimbrial Adhesins of Enterotoxigenic Escherichia coli
Biology (Basel), 2, 2013
|
|
4K6T
 
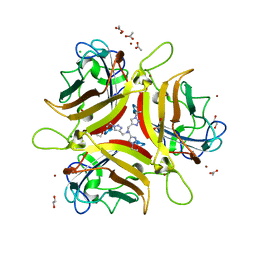 | | Crystal structure of Ad37 fiber knob in complex with trivalent sialic acid inhibitor ME0385 | | Descriptor: | 1,2-ETHANEDIOL, 2,2',2''-[nitrilotris(methanediyl-1H-1,2,3-triazole-4,1-diyl)]triethanol, ACETATE ION, ... | | Authors: | Stehle, T, Bauer, J. | | Deposit date: | 2013-04-16 | | Release date: | 2014-10-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Triazole linker-based trivalent sialic acid inhibitors of adenovirus type 37 infection of human corneal epithelial cells.
Org.Biomol.Chem., 13, 2015
|
|
4KNN
 
 | | Crystal structure of human carbonic anhydrase isozyme XIII with 2-Chloro-4-[(pyrimidin-2-ylsulfanyl)acetyl]benzenesulfonamide | | Descriptor: | 1,2-ETHANEDIOL, 2-chloro-4-[(pyrimidin-2-ylsulfanyl)acetyl]benzenesulfonamide, ACETIC ACID, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2013-05-10 | | Release date: | 2013-11-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.404 Å) | | Cite: | Benzenesulfonamides with pyrimidine moiety as inhibitors of human carbonic anhydrases I, II, VI, VII, XII, and XIII
Bioorg.Med.Chem., 21, 2013
|
|
4KFN
 
 | | Structure-Based Discovery of Novel Amide-Containing Nicotinamide Phosphoribosyltransferase (Nampt) Inhibitors | | Descriptor: | 1,2-ETHANEDIOL, N-[4-(piperidin-1-ylsulfonyl)benzyl]-1H-pyrrolo[3,2-c]pyridine-2-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Zheng, X, Bauer, P, Baumeister, T, Buckmelter, A.J, Caligiuri, M, Clodfelter, K.H, Han, B, Ho, Y, Kley, N, Lin, J, Reynolds, D.J, Sharma, G, Smith, C.C, Wang, Z, Dragovich, P.S, Gunzner-Toste, J, Liederer, B.M, Ly, J, O'Brien, T, Oh, A, Wang, L, Wang, W, Xiao, Y, Zak, M, Zhao, G, Yuen, P, Bair, K.W. | | Deposit date: | 2013-04-27 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-based identification of ureas as novel nicotinamide phosphoribosyltransferase (nampt) inhibitors.
J.Med.Chem., 56, 2013
|
|
4KH8
 
 | |
4KWY
 
 | |
4KW2
 
 | |
4LER
 
 | |
4KZ2
 
 | | Crystal Structure of phi29 pRNA 3WJ Core | | Descriptor: | MANGANESE (II) ION, phi29 pRNA 3WJ core RNA 16 mer, phi29 pRNA 3WJ core RNA 18 mer, ... | | Authors: | Zhang, H, Endrizzi, J.A, Shu, Y, Haque, F, Sauter, C, Guo, P, Chi, Y.-I. | | Deposit date: | 2013-05-29 | | Release date: | 2013-10-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal structure of 3WJ core revealing divalent ion-promoted thermostability and assembly of the Phi29 hexameric motor pRNA.
Rna, 19, 2013
|
|
4L1N
 
 | |
