6RJJ
 
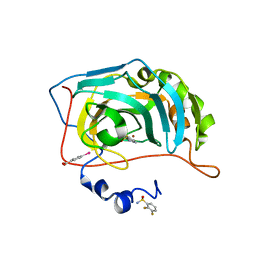 | | Human Carbonic Anhydrase II in complex with fluorinated benzenesulfonamide | | Descriptor: | (4-CARBOXYPHENYL)(CHLORO)MERCURY, 2,3-bis(fluoranyl)benzenesulfonamide, Carbonic anhydrase 2, ... | | Authors: | Gloeckner, S, Heine, A, Klebe, G. | | Deposit date: | 2019-04-27 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.056 Å) | | Cite: | The Influence of Varying Fluorination Patterns on the Thermodynamics and Kinetics of Benzenesulfonamide Binding to Human Carbonic Anhydrase II.
Biomolecules, 10, 2020
|
|
6RQI
 
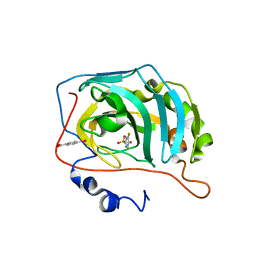 | | Human Carbonic Anhydrase II in complex with fluorinated benzenesulfonamide | | Descriptor: | (4-CARBOXYPHENYL)(CHLORO)MERCURY, 3-fluorobenzenesulfonamide, Carbonic anhydrase 2, ... | | Authors: | Gloeckner, S, Heine, A, Klebe, G. | | Deposit date: | 2019-05-15 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | The Influence of Varying Fluorination Patterns on the Thermodynamics and Kinetics of Benzenesulfonamide Binding to Human Carbonic Anhydrase II.
Biomolecules, 10, 2020
|
|
8IPT
 
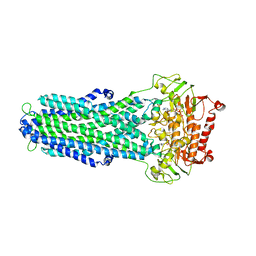 | |
8IPS
 
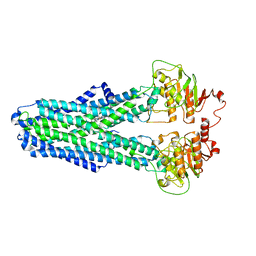 | |
8IPR
 
 | |
8G8C
 
 | | Crystal structure of DH1322.1 Fab in complex with HIV proximal MPER peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DH1322.1 heavy chain, DH1322.1 light chain, ... | | Authors: | Niyongabo, A, Janus, B.M, Ofek, G. | | Deposit date: | 2023-02-17 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Vaccine induction of heterologous HIV-1-neutralizing antibody B cell lineages in humans.
Cell, 187, 2024
|
|
8VRE
 
 | | Structure of PYCR1 complexed with NADH and N-formyl-L-proline | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 1-formyl-L-proline, Pyrroline-5-carboxylate reductase 1, ... | | Authors: | Tanner, J.J, Meeks, K.R. | | Deposit date: | 2024-01-21 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Screening a knowledge-based library of low molecular weight compounds against the proline biosynthetic enzyme 1-pyrroline-5-carboxylate 1 (PYCR1)
Protein Sci., 33, 2024
|
|
6RKN
 
 | | Human Carbonic Anhydrase II in complex with a fluorinated Benzenesulfonamide. | | Descriptor: | (4-CARBOXYPHENYL)(CHLORO)MERCURY, 4-FLUOROBENZENESULFONAMIDE, Carbonic anhydrase 2, ... | | Authors: | Gloeckner, S, Heine, A, Klebe, G. | | Deposit date: | 2019-04-30 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.959 Å) | | Cite: | The Influence of Varying Fluorination Patterns on the Thermodynamics and Kinetics of Benzenesulfonamide Binding to Human Carbonic Anhydrase II.
Biomolecules, 10, 2020
|
|
3LIO
 
 | | X-ray structure of the iron superoxide dismutase from pseudoalteromonas haloplanktis (crystal form I) | | Descriptor: | FE (III) ION, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose, iron superoxide dismutase | | Authors: | Merlino, A, Russo Krauss, I, Rossi, B, Conte, M, Vergara, A, Sica, F. | | Deposit date: | 2010-01-25 | | Release date: | 2010-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and flexibility in cold-adapted iron superoxide dismutases: the case of the enzyme isolated from Pseudoalteromonas haloplanktis.
J.Struct.Biol., 172, 2010
|
|
3MBA
 
 | | APLYSIA LIMACINA MYOGLOBIN. CRYSTALLOGRAPHIC ANALYSIS AT 1.6 ANGSTROMS RESOLUTION | | Descriptor: | FLUORIDE ION, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bolognesi, M, Onesti, S, Gatti, G, Coda, A, Ascenzi, P, Brunori, M. | | Deposit date: | 1989-02-22 | | Release date: | 1990-01-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Aplysia limacina myoglobin. Crystallographic analysis at 1.6 A resolution.
J.Mol.Biol., 205, 1989
|
|
6RNP
 
 | | Human Carbonic Anhydrase II in complex with fluorinated benzenesulfonamide | | Descriptor: | (4-CARBOXYPHENYL)(CHLORO)MERCURY, 2,5-bis(fluoranyl)benzenesulfonamide, Carbonic anhydrase 2, ... | | Authors: | Gloeckner, S, Heine, A, Klebe, G. | | Deposit date: | 2019-05-09 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | The Influence of Varying Fluorination Patterns on the Thermodynamics and Kinetics of Benzenesulfonamide Binding to Human Carbonic Anhydrase II.
Biomolecules, 10, 2020
|
|
6ROE
 
 | | Human Carbonic Anhydrase II in complex with fluorinated benzenesulfonamide | | Descriptor: | (4-CARBOXYPHENYL)(CHLORO)MERCURY, 2,6-DIFLUOROBENZENESULFONAMIDE, Carbonic anhydrase 2, ... | | Authors: | Gloeckner, S, Heine, A, Klebe, G. | | Deposit date: | 2019-05-12 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.939 Å) | | Cite: | The Influence of Varying Fluorination Patterns on the Thermodynamics and Kinetics of Benzenesulfonamide Binding to Human Carbonic Anhydrase II.
Biomolecules, 10, 2020
|
|
2BTW
 
 | | Crystal structure of Alr0975 | | Descriptor: | ALR0975 PROTEIN, CALCIUM ION | | Authors: | Vivares, D, Arnoux, P, Pignol, D. | | Deposit date: | 2005-06-07 | | Release date: | 2005-12-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Papain-Like Enzyme at Work: Native and Acyl- Enzyme Intermediate Structures in Phytochelatin Synthesis.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
3FL3
 
 | | X-ray structure of the ligand free non covalent swapped form of the A19P/Q28L/K31C/S32C mutant of bovine pancreatic ribonuclease | | Descriptor: | Ribonuclease pancreatic, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose | | Authors: | Merlino, A, Russo Krauss, I, Perillo, M, Mattia, C.A, Ercole, C, Picone, D, Vergara, A, Sica, F. | | Deposit date: | 2008-12-18 | | Release date: | 2009-03-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Toward an antitumor form of bovine pancreatic ribonuclease: The crystal structure of three noncovalent dimeric mutants
Biopolymers, 91, 2009
|
|
5G1P
 
 | | Aspartate transcarbamoylase domain of human CAD bound to carbamoyl phosphate | | Descriptor: | CAD PROTEIN, PHOSPHORIC ACID MONO(FORMAMIDE)ESTER | | Authors: | Ruiz-Ramos, A, Grande-Garcia, A, Moreno-Morcillo, M.D, Ramon-Maiques, S. | | Deposit date: | 2016-03-29 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structure and Functional Characterization of Human Aspartate Transcarbamoylase, the Target of the Anti-Tumoral Drug Pala.
Structure, 24, 2016
|
|
1V6M
 
 | | Peanut Lectin with 9mer peptide (IWSSAGNVA) | | Descriptor: | CALCIUM ION, Galactose-binding lectin, MANGANESE (II) ION | | Authors: | Kundhavai Natchiar, S, Arockia Jeyaprakash, A, Ramya, T.N.C, Thomas, C.J, Suguna, K, Surolia, A, Vijayan, M. | | Deposit date: | 2003-12-02 | | Release date: | 2004-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural plasticity of peanut lectin: an X-ray analysis involving variation in pH, ligand binding and crystal structure.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
3FL0
 
 | | X-ray structure of the non covalent swapped form of the Q28L/K31C/S32C mutant of bovine pancreatic ribonuclease in complex with 2'-DEOXYCYTIDINE-2'-DEOXYGUANOSINE-3',5'-MONOPHOSPHATE | | Descriptor: | 2'-DEOXYCYTIDINE-2'-DEOXYGUANOSINE-3',5'-MONOPHOSPHATE, Ribonuclease pancreatic | | Authors: | Merlino, A, Russo Krauss, I, Perillo, M, Mattia, C.A, Ercole, C, Picone, D, Vergara, A, Sica, F. | | Deposit date: | 2008-12-18 | | Release date: | 2009-03-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Toward an antitumor form of bovine pancreatic ribonuclease: The crystal structure of three noncovalent dimeric mutants
Biopolymers, 91, 2009
|
|
6RL9
 
 | | Human Carbonic Anhydrase II in complex with 4-Aminobenzenesulfonamide | | Descriptor: | (4-CARBOXYPHENYL)(CHLORO)MERCURY, Carbonic anhydrase 2, MERCURY (II) ION, ... | | Authors: | Gloeckner, S, Heine, A, Klebe, G. | | Deposit date: | 2019-05-01 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | The Influence of Varying Fluorination Patterns on the Thermodynamics and Kinetics of Benzenesulfonamide Binding to Human Carbonic Anhydrase II.
Biomolecules, 10, 2020
|
|
6RSZ
 
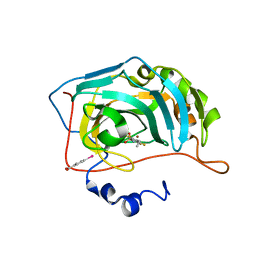 | | Human Carbonic Anhydrase II in complex with fluorinated benzenesulfonamide | | Descriptor: | (4-CARBOXYPHENYL)(CHLORO)MERCURY, 4-ethyl-2,3,5,6-tetrakis(fluoranyl)benzenesulfonamide, Carbonic anhydrase 2, ... | | Authors: | Gloeckner, S, Ngo, K, Heine, A, Klebe, G. | | Deposit date: | 2019-05-22 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.089 Å) | | Cite: | The Influence of Varying Fluorination Patterns on the Thermodynamics and Kinetics of Benzenesulfonamide Binding to Human Carbonic Anhydrase II.
Biomolecules, 10, 2020
|
|
5I4R
 
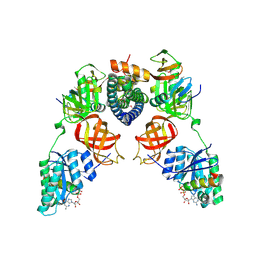 | | Contact-dependent inhibition system from Escherichia coli NC101 - ternary CdiA/CdiI/EF-Tu complex (trypsin-modified) | | Descriptor: | Contact-dependent inhibitor A, Contact-dependent inhibitor I, Elongation factor Tu, ... | | Authors: | Michalska, K, Stols, L, Eschenfeldt, W, Hayes, C.S, Goulding, C.W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI) | | Deposit date: | 2016-02-12 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of a novel antibacterial toxin that exploits elongation factor Tu to cleave specific transfer RNAs.
Nucleic Acids Res., 45, 2017
|
|
3FL1
 
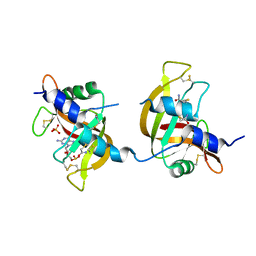 | | X-ray structure of the non covalent swapped form of the A19P/Q28L/K31C/S32C mutant of bovine pancreatic ribonuclease in complex with 2'-DEOXYCYTIDINE-2'-DEOXYGUANOSINE-3',5'-MONOPHOSPHATE | | Descriptor: | 2'-DEOXYCYTIDINE-2'-DEOXYGUANOSINE-3',5'-MONOPHOSPHATE, Ribonuclease pancreatic, SULFATE ION, ... | | Authors: | Merlino, A, Russo Krauss, I, Perillo, M, Mattia, C.A, Ercole, C, Picone, D, Vergara, A, Sica, F. | | Deposit date: | 2008-12-18 | | Release date: | 2009-03-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Toward an antitumor form of bovine pancreatic ribonuclease: The crystal structure of three noncovalent dimeric mutants
Biopolymers, 91, 2009
|
|
6M1H
 
 | | CryoEM structure of human PAC1 receptor in complex with maxadilan | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Song, X, Wang, J, Zhang, D, Wang, H.W, Ma, Y. | | Deposit date: | 2020-02-26 | | Release date: | 2020-03-11 | | Last modified: | 2020-05-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures of PAC1 receptor reveal ligand binding mechanism.
Cell Res., 30, 2020
|
|
1T7D
 
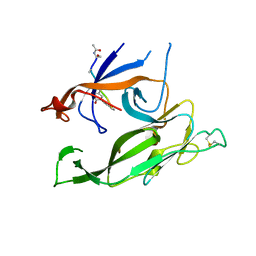 | | Crystal structure of Escherichia coli type I signal peptidase in complex with a lipopeptide inhibitor | | Descriptor: | 10-METHYLUNDECANOIC ACID, ARYLOMYCIN A2, SIGNAL PEPTIDASE I | | Authors: | Paetzel, M, Goodall, J.J, Kania, M, Dalbey, R.E, Page, M.G.P. | | Deposit date: | 2004-05-09 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystallographic and Biophysical Analysis of a Bacterial Signal Peptidase in Complex with a Lipopeptide Based Inhibitor.
J.Biol.Chem., 279, 2004
|
|
6RRI
 
 | | Human Carbonic Anhydrase II in complex with fluorinated benzenesulfonamide | | Descriptor: | (4-CARBOXYPHENYL)(CHLORO)MERCURY, 2,3,5,6-tetrakis(fluoranyl)benzenesulfonamide, Carbonic anhydrase 2, ... | | Authors: | Gloeckner, S, Heine, A, Klebe, G. | | Deposit date: | 2019-05-18 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.097 Å) | | Cite: | The Influence of Varying Fluorination Patterns on the Thermodynamics and Kinetics of Benzenesulfonamide Binding to Human Carbonic Anhydrase II.
Biomolecules, 10, 2020
|
|
1I8P
 
 | |
