1IA9
 
 | | CRYSTAL STRUCTURE OF THE ATYPICAL PROTEIN KINASE DOMAIN OF A TRP CA-CHANNEL, CHAK (AMPPNP COMPLEX) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, TRANSIENT RECEPTOR POTENTIAL-RELATED PROTEIN, ... | | Authors: | Yamaguchi, H, Matsushita, M, Nairn, A.C, Kuriyan, J. | | Deposit date: | 2001-03-22 | | Release date: | 2001-06-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the atypical protein kinase domain of a TRP channel with phosphotransferase activity.
Mol.Cell, 7, 2001
|
|
1XQS
 
 | | Crystal structure of the HspBP1 core domain complexed with the fragment of Hsp70 ATPase domain | | Descriptor: | ADENOSINE MONOPHOSPHATE, HSPBP1 protein, Heat shock 70 kDa protein 1 | | Authors: | Shomura, Y, Dragovic, Z, Chang, H.C, Tzvetkov, N, Young, J.C, Brodsky, J.L, Guerriero, V, Hartl, F.U, Bracher, A. | | Deposit date: | 2004-10-13 | | Release date: | 2005-03-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Regulation of Hsp70 Function by HspBP1; Structural Analysis Reveals an Alternate Mechanism for Hsp70 Nucleotide Exchange
Mol.Cell, 17, 2005
|
|
2DZC
 
 | | Crystal Structure Of Biotin Protein Ligase From Pyrococcus Horikoshii, Mutation R48A | | Descriptor: | biotin--[acetyl-CoA-carboxylase] ligase | | Authors: | Bagautdinov, B, Taketa, M, Matsuura, Y, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-09-27 | | Release date: | 2007-03-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Protein biotinylation visualized by a complex structure of biotin protein ligase with a substrate
J.Biol.Chem., 283, 2008
|
|
1FRN
 
 | |
1KI9
 
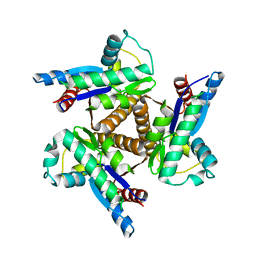 | |
2F1I
 
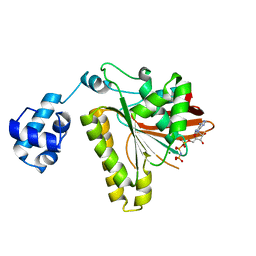 | | Recombinase in Complex with AMP-PNP | | Descriptor: | DNA repair and recombination protein radA, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Qian, X, He, Y, Wu, Y, Luo, Y. | | Deposit date: | 2005-11-14 | | Release date: | 2006-05-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Asp302 determines potassium dependence of a RadA recombinase from Methanococcus voltae.
J.Mol.Biol., 360, 2006
|
|
2DEJ
 
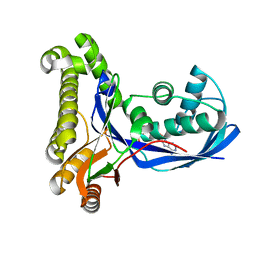 | | Crystal Structure of galaktokinase from Pyrococcus horikoshii with AMP-PN and galactose | | Descriptor: | Probable galactokinase, alpha-D-galactopyranose, {5'-O-[(R)-{[(S)-AMINO(HYDROXY-KAPPAO)PHOSPHORYL]OXY}(HYDROXY-KAPPAO)PHOSPHORYL]ADENOSINATO(2-)}MAGNESIUM | | Authors: | Inagaki, E, Sakamoto, K, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-02-10 | | Release date: | 2007-05-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of galaktokinase from Pyrococcus horikoshii
To be Published
|
|
3GDE
 
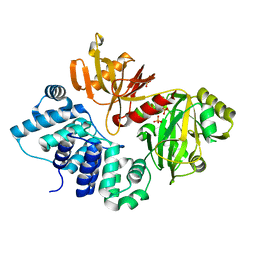 | | The closed conformation of ATP-dependent DNA ligase from Archaeoglobus fulgidus | | Descriptor: | DNA ligase, PHOSPHATE ION | | Authors: | Kim, D.J, Kim, H.-W, Kim, O, Kim, H.S, Lee, S.J, Suh, S.W. | | Deposit date: | 2009-02-24 | | Release date: | 2009-12-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | ATP-dependent DNA ligase from Archaeoglobus fulgidus displays a tightly closed conformation
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3AGO
 
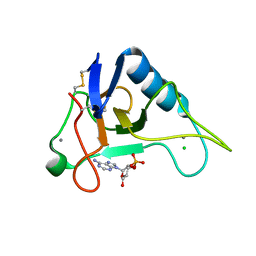 | |
1COW
 
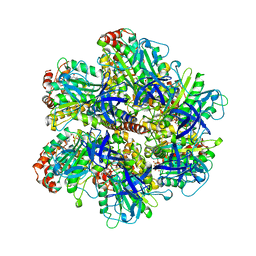 | | BOVINE MITOCHONDRIAL F1-ATPASE COMPLEXED WITH AUROVERTIN B | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, AUROVERTIN B, BOVINE MITOCHONDRIAL F1-ATPASE, ... | | Authors: | van Raaij, M.J, Abrahams, J.P, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 1996-05-08 | | Release date: | 1996-08-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structure of bovine F1-ATPase complexed with the antibiotic inhibitor aurovertin B.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
2HVQ
 
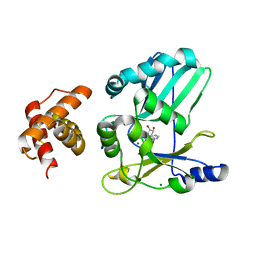 | | Structure of Adenylated full-length T4 RNA Ligase 2 | | Descriptor: | Hypothetical 37.6 kDa protein in Gp24-hoc intergenic region, MAGNESIUM ION | | Authors: | Nandakumar, J, Lima, C.D. | | Deposit date: | 2006-07-30 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | RNA Ligase Structures Reveal the Basis for RNA Specificity and Conformational Changes that Drive Ligation Forward.
Cell(Cambridge,Mass.), 127, 2006
|
|
2HVR
 
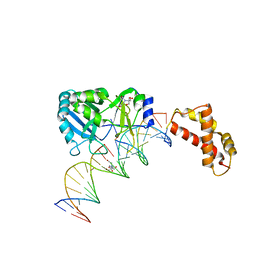 | | Structure of T4 RNA Ligase 2 with Nicked 5'-Adenylated nucleic acid duplex containing a 3'-deoxyribonucleotide at the nick | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-D(*AP*TP*TP*CP*CP*GP*AP*TP*AP*GP*TP*GP*GP*GP*GP*TP*CP*GP*CP*AP*AP*TP*TP*G)-3', 5'-D(*CP*AP*AP*TP*TP*GP*CP*GP*AP*C)-R(P*(OMC)P*C)-3', ... | | Authors: | Nandakumar, J, Lima, C.D. | | Deposit date: | 2006-07-30 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | RNA Ligase Structures Reveal the Basis for RNA Specificity and Conformational Changes that Drive Ligation Forward.
Cell(Cambridge,Mass.), 127, 2006
|
|
2DGN
 
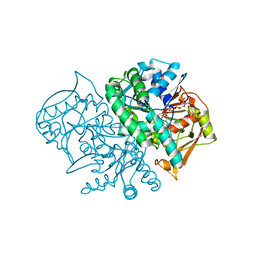 | | Mouse Muscle Adenylosuccinate Synthetase partially ligated complex with GTP, 2'-deoxy-IMP | | Descriptor: | 9-(2-DEOXY-5-O-PHOSPHONO-BETA-D-ERYTHRO-PENTOFURANOSYL)-6-(PHOSPHONOOXY)-9H-PURINE, Adenylosuccinate synthetase isozyme 1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Iancu, C.V, Zhou, Y, Borza, T, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2006-03-15 | | Release date: | 2006-09-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cavitation as a mechanism of substrate discrimination by adenylosuccinate synthetases.
Biochemistry, 45, 2006
|
|
7PD9
 
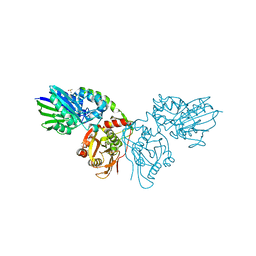 | |
7PCP
 
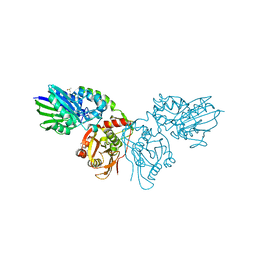 | |
2AKY
 
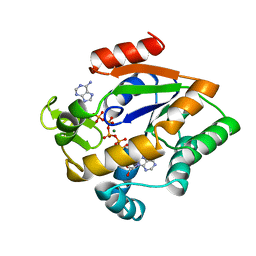 | |
8JGK
 
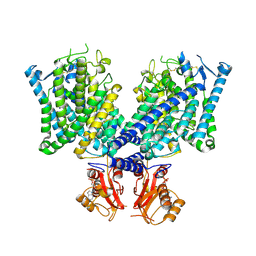 | | Cryo-EM structure of mClC-3 with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, H(+)/Cl(-) exchange transporter 3 | | Authors: | Wan, Y.Z.Q, Yang, F. | | Deposit date: | 2023-05-21 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (4.04 Å) | | Cite: | Structural basis of adenine nucleotides regulation and neurodegenerative pathology in ClC-3 exchanger.
Nat Commun, 15, 2024
|
|
8JGS
 
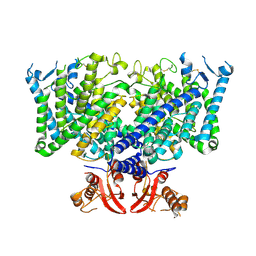 | | Cryo-EM structure of apo state mClC-3_I607T | | Descriptor: | H(+)/Cl(-) exchange transporter 3 | | Authors: | Wan, Y.Z.Q, Yang, F. | | Deposit date: | 2023-05-21 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Structural basis of adenine nucleotides regulation and neurodegenerative pathology in ClC-3 exchanger.
Nat Commun, 15, 2024
|
|
8JGJ
 
 | | Cryo-EM structure of mClC-3 with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, H(+)/Cl(-) exchange transporter 3 | | Authors: | Wan, Y.Z.Q, Yang, F. | | Deposit date: | 2023-05-20 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of adenine nucleotides regulation and neurodegenerative pathology in ClC-3 exchanger.
Nat Commun, 15, 2024
|
|
8JGV
 
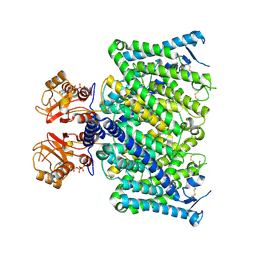 | | Cryo-EM structure of mClC-3_I607T with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, H(+)/Cl(-) exchange transporter 3 | | Authors: | Wan, Y.Z.Q, Yang, F. | | Deposit date: | 2023-05-21 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural basis of adenine nucleotides regulation and neurodegenerative pathology in ClC-3 exchanger.
Nat Commun, 15, 2024
|
|
2JDV
 
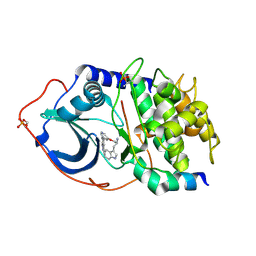 | | Structure of PKA-PKB chimera complexed with A-443654 | | Descriptor: | (2S)-1-(1H-INDOL-3-YL)-3-{[5-(3-METHYL-1H-INDAZOL-5-YL)PYRIDIN-3-YL]OXY}PROPAN-2-AMINE, CAMP-DEPENDENT PROTEIN KINASE, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA | | Authors: | Davies, T.G, Verdonk, M.L, Graham, B, Saalau-Bethell, S, Hamlett, C.C.F, Mchardy, T, Collins, I, Garrett, M.D, Workman, P, Woodhead, S.J, Jhoti, H, Barford, D. | | Deposit date: | 2007-01-12 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | A Structural Comparison of Inhibitor Binding to Pkb, Pka and Pka-Pkb Chimera
J.Mol.Biol., 367, 2007
|
|
7TDK
 
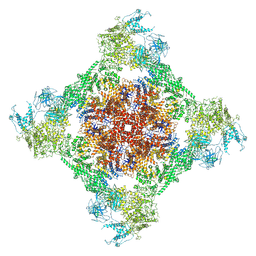 | | Rabbit RyR1 with AMP-PCP and high Ca2+ embedded in nanodisc in closed-inactivated conformation class 3 (Dataset-A) | | Descriptor: | CALCIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Ryanodine receptor 1,Ryanodine receptor 1,RyR1, ... | | Authors: | Nayak, A.R, Samso, M. | | Deposit date: | 2021-12-31 | | Release date: | 2022-03-09 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Ca 2+ -inactivation of the mammalian ryanodine receptor type 1 in a lipidic environment revealed by cryo-EM.
Elife, 11, 2022
|
|
7TDH
 
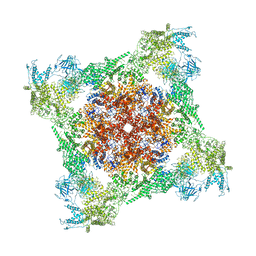 | | Rabbit RyR1 with AMP-PCP and high Ca2+ embedded in nanodisc in open conformation | | Descriptor: | CALCIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Ryanodine receptor 1,Ryanodine receptor 1,RyR1, ... | | Authors: | Nayak, A.R, Samso, M. | | Deposit date: | 2021-12-31 | | Release date: | 2022-03-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Ca 2+ -inactivation of the mammalian ryanodine receptor type 1 in a lipidic environment revealed by cryo-EM.
Elife, 11, 2022
|
|
7TDI
 
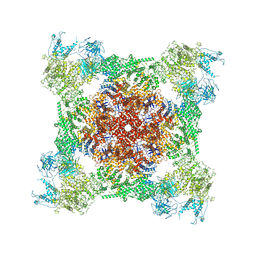 | | Rabbit RyR1 with AMP-PCP and high Ca2+ embedded in nanodisc in closed-inactivated conformation class 2 (Dataset-A) | | Descriptor: | CALCIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Ryanodine receptor 1,Ryanodine receptor 1,RyR1, ... | | Authors: | Nayak, A.R, Samso, M. | | Deposit date: | 2021-12-31 | | Release date: | 2022-03-09 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Ca 2+ -inactivation of the mammalian ryanodine receptor type 1 in a lipidic environment revealed by cryo-EM.
Elife, 11, 2022
|
|
8JEV
 
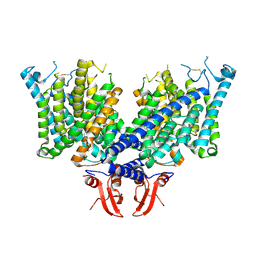 | | Cryo-EM structure of apo state mClC-3 | | Descriptor: | CHLORIDE ION, H(+)/Cl(-) exchange transporter 3 | | Authors: | Wan, Y.Z.Q, Yang, F. | | Deposit date: | 2023-05-16 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural basis of adenine nucleotides regulation and neurodegenerative pathology in ClC-3 exchanger.
Nat Commun, 15, 2024
|
|
