7YUM
 
 | | MtaLon-Apo for the spiral oligomers of tetramer | | Descriptor: | Lon protease | | Authors: | Li, S, Hsieh, K, Kuo, C, Lee, S, Ho, M, Wang, C, Zhang, K, Chang, C.I. | | Deposit date: | 2022-08-17 | | Release date: | 2023-10-25 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
7YUX
 
 | | MtaLon-ADP for the spiral oligomers of hexamer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease | | Authors: | Li, S, Hsieh, K, Kuo, C, Lee, S, Ho, M, Wang, C, Zhang, K, Chang, C.I. | | Deposit date: | 2022-08-18 | | Release date: | 2023-10-25 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
8JUW
 
 | | Human ATAD2 Walker B mutant-H3/H4K5Q complex, ATP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATPase family AAA domain-containing protein 2 | | Authors: | Cho, C, Song, J. | | Deposit date: | 2023-06-27 | | Release date: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Structure of the human ATAD2 AAA+ histone chaperone reveals mechanism of regulation and inter-subunit communication.
Commun Biol, 6, 2023
|
|
8JUZ
 
 | | Human ATAD2 Walker B mutant-H3/H4K5Q complex, ATP state (Class III) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATPase family AAA domain-containing protein 2 | | Authors: | Cho, C, Song, J. | | Deposit date: | 2023-06-27 | | Release date: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (4.29 Å) | | Cite: | Structure of the human ATAD2 AAA+ histone chaperone reveals mechanism of regulation and inter-subunit communication.
Commun Biol, 6, 2023
|
|
8JUY
 
 | | Human ATAD2 Walker B mutant-H3/H4K5Q complex, ATP state (Class II) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATPase family AAA domain-containing protein 2 | | Authors: | Cho, C, Song, J. | | Deposit date: | 2023-06-27 | | Release date: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (4.34 Å) | | Cite: | Structure of the human ATAD2 AAA+ histone chaperone reveals mechanism of regulation and inter-subunit communication.
Commun Biol, 6, 2023
|
|
7Y53
 
 | | The cryo-EM structure of human ERAD retro-translocation complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Derlin-1, Transitional endoplasmic reticulum ATPase | | Authors: | Cao, Y, Rao, B, Wang, Q, Yao, D, Xia, Y, Li, W, Li, S, Shen, Y. | | Deposit date: | 2022-06-16 | | Release date: | 2023-10-18 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | The cryo-EM structure of the human ERAD retrotranslocation complex.
Sci Adv, 9, 2023
|
|
7Y59
 
 | | The cryo-EM structure of human ERAD retro-translocation complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Derlin-1, ... | | Authors: | Cao, Y, Rao, B, Wang, Q, Yao, D, Xia, Y, Li, W, Li, S, Shen, Y. | | Deposit date: | 2022-06-16 | | Release date: | 2023-10-18 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (4.51 Å) | | Cite: | The cryo-EM structure of the human ERAD retrotranslocation complex.
Sci Adv, 9, 2023
|
|
7Y4W
 
 | | The cryo-EM structure of human ERAD retro-translocation complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Derlin-1, Transitional endoplasmic reticulum ATPase | | Authors: | Cao, Y, Rao, B, Wang, Q, Yao, D, Xia, Y, Li, W, Li, S, Shen, Y. | | Deposit date: | 2022-06-16 | | Release date: | 2023-10-18 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | The cryo-EM structure of the human ERAD retrotranslocation complex.
Sci Adv, 9, 2023
|
|
8H3H
 
 | | Human ATAD2 Walker B mutant, ATP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATPase family AAA domain-containing protein 2 | | Authors: | Cho, C, Song, J. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structure of the human ATAD2 AAA+ histone chaperone reveals mechanism of regulation and inter-subunit communication.
Commun Biol, 6, 2023
|
|
8C0W
 
 | | Structure of the peroxisomal Pex1/Pex6 ATPase complex bound to a substrate in twin seam state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ruettermann, M, Koci, M, Lill, P, Geladas, E.D, Kaschani, F, Klink, B.U, Erdmann, R, Gatsogiannis, C. | | Deposit date: | 2022-12-19 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structure of the peroxisomal Pex1/Pex6 ATPase complex bound to a substrate.
Nat Commun, 14, 2023
|
|
8C0V
 
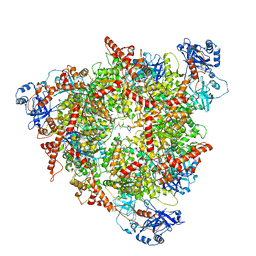 | | Structure of the peroxisomal Pex1/Pex6 ATPase complex bound to a substrate in single seam state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ruettermann, M, Koci, M, Lill, P, Geladas, E.D, Kaschani, F, Klink, B.U, Erdmann, R, Gatsogiannis, C. | | Deposit date: | 2022-12-19 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of the peroxisomal Pex1/Pex6 ATPase complex bound to a substrate.
Nat Commun, 14, 2023
|
|
8TID
 
 | | Combined linker domain of N-DRC and associated proteins Tetrahymena | | Descriptor: | AAA family ATPase CDC48 subfamily protein, CFAP20, Calmodulin 7-2, ... | | Authors: | Ghanaeian, A.G, Majhi, S.M, McCaffrey, C.M, Nami, B.N, Black, C.B, Yang, S.K, Legal, T.L, Papoulas, O.P, Janowska, M.J, Valente-Paterno, M.V, Marcotte, E.M, Wloga, D.W, Bui, K.H. | | Deposit date: | 2023-07-19 | | Release date: | 2023-09-27 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Integrated modeling of the Nexin-dynein regulatory complex reveals its regulatory mechanism.
Nat Commun, 14, 2023
|
|
8TH8
 
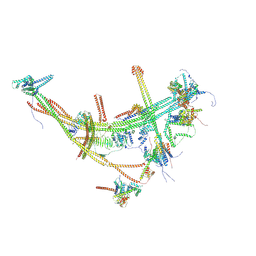 | |
8J07
 
 | |
8HL7
 
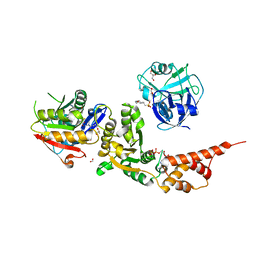 | |
8FCO
 
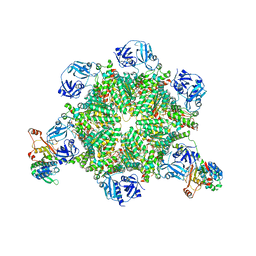 | | Cryo-EM structure of p97:UBXD1 meta state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase, UBX domain-containing protein 6 | | Authors: | Braxton, J.R, Tucker, M.R, Tse, E, Southworth, D.R. | | Deposit date: | 2022-12-01 | | Release date: | 2023-06-21 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | The p97/VCP adaptor UBXD1 drives AAA+ remodeling and ring opening through multi-domain tethered interactions.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8FCQ
 
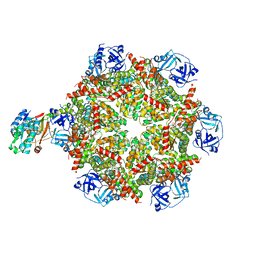 | | Cryo-EM structure of p97:UBXD1 PUB-in state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase, UBX domain-containing protein 6 | | Authors: | Braxton, J.R, Tucker, M.R, Tse, E, Southworth, D.R. | | Deposit date: | 2022-12-01 | | Release date: | 2023-06-21 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | The p97/VCP adaptor UBXD1 drives AAA+ remodeling and ring opening through multi-domain tethered interactions.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8FCM
 
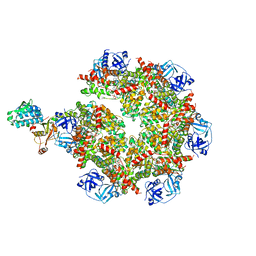 | | Cryo-EM structure of p97:UBXD1 open state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase, UBX domain-containing protein 6 | | Authors: | Braxton, J.R, Tucker, M.R, Tse, E, Southworth, D.R. | | Deposit date: | 2022-12-01 | | Release date: | 2023-06-21 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | The p97/VCP adaptor UBXD1 drives AAA+ remodeling and ring opening through multi-domain tethered interactions.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8FCN
 
 | | Cryo-EM structure of p97:UBXD1 VIM-only state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase, UBX domain-containing protein 6 | | Authors: | Braxton, J.R, Tucker, M.R, Tse, E, Southworth, D.R. | | Deposit date: | 2022-12-01 | | Release date: | 2023-06-21 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | The p97/VCP adaptor UBXD1 drives AAA+ remodeling and ring opening through multi-domain tethered interactions.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8FCT
 
 | | Cryo-EM structure of p97:UBXD1 lariat mutant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase, UBX domain-containing protein 6 | | Authors: | Braxton, J.R, Tucker, M.R, Tse, E, Southworth, D.R. | | Deposit date: | 2022-12-01 | | Release date: | 2023-06-21 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | The p97/VCP adaptor UBXD1 drives AAA+ remodeling and ring opening through multi-domain tethered interactions.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8FCL
 
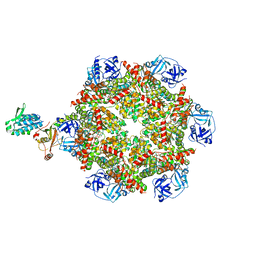 | | Cryo-EM structure of p97:UBXD1 closed state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase, UBX domain-containing protein 6 | | Authors: | Braxton, J.R, Tucker, M.R, Tse, E, Southworth, D.R. | | Deposit date: | 2022-12-01 | | Release date: | 2023-06-21 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | The p97/VCP adaptor UBXD1 drives AAA+ remodeling and ring opening through multi-domain tethered interactions.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8FCP
 
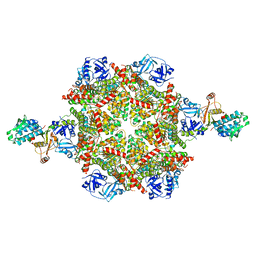 | | Cryo-EM structure of p97:UBXD1 para state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase, UBX domain-containing protein 6 | | Authors: | Braxton, J.R, Tucker, M.R, Tse, E, Southworth, D.R. | | Deposit date: | 2022-12-01 | | Release date: | 2023-06-21 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | The p97/VCP adaptor UBXD1 drives AAA+ remodeling and ring opening through multi-domain tethered interactions.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8FCR
 
 | | Cryo-EM structure of p97:UBXD1 H4-bound state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase, UBX domain-containing protein 6 | | Authors: | Braxton, J.R, Tucker, M.R, Tse, E, Southworth, D.R. | | Deposit date: | 2022-12-01 | | Release date: | 2023-06-21 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (4.12 Å) | | Cite: | The p97/VCP adaptor UBXD1 drives AAA+ remodeling and ring opening through multi-domain tethered interactions.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8FS4
 
 | | Structure of S. cerevisiae Rad24-RFC loading the 9-1-1 clamp onto a 10-nt gapped DNA in step 2 (open 9-1-1 ring and flexibly bound chamber DNA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, DDC1 isoform 1, ... | | Authors: | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | Deposit date: | 2023-01-09 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structures of 9-1-1 DNA checkpoint clamp loading at gaps from start to finish and ramification to biology.
Biorxiv, 2023
|
|
8FS5
 
 | | Structure of S. cerevisiae Rad24-RFC loading the 9-1-1 clamp onto a 10-nt gapped DNA in step 3 (open 9-1-1 and stably bound chamber DNA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, DDC1 isoform 1, ... | | Authors: | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | Deposit date: | 2023-01-09 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structures of 9-1-1 DNA checkpoint clamp loading at gaps from start to finish and ramification to biology.
Biorxiv, 2023
|
|
