9C83
 
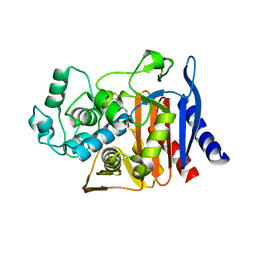 | | X-ray crystal structure of AmpC beta-lactamase with inhibitor | | Descriptor: | AmpC Beta-lactamase, N-[(3M)-3-(5-chloro-1,2,3-thiadiazol-4-yl)phenyl]-5-methyl-3-oxo-2,3-dihydro-1,2-oxazole-4-sulfonamide | | Authors: | Liu, F, Shoichet, B.K. | | Deposit date: | 2024-06-11 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Improved correlations with score, hit-rate, and affinity as docking library and testing scale increase
To Be Published
|
|
1FB5
 
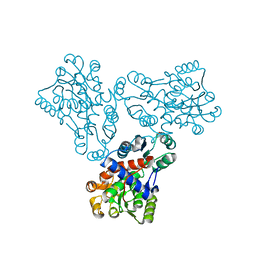 | | LOW RESOLUTION STRUCTURE OF OVINE ORNITHINE TRANSCARBMOYLASE IN THE UNLIGANDED STATE | | Descriptor: | NORVALINE, ORNITHINE TRANSCARBAMOYLASE | | Authors: | Zanotti, G, Battistutta, R, Panzalorto, M, Francescato, P, Bruno, G, De Gregorio, A. | | Deposit date: | 2000-07-14 | | Release date: | 2003-08-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Functional and structural characterization of ovine ornithine transcarbamoylase.
Org.Biomol.Chem., 1, 2003
|
|
2AIJ
 
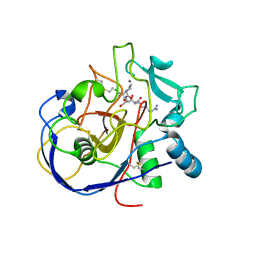 | |
9C84
 
 | |
2AIK
 
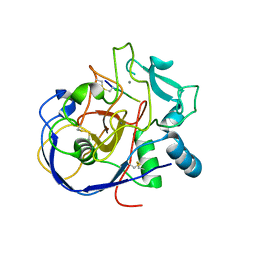 | |
1CS1
 
 | |
4E1V
 
 | | X-RAY Structure of the Uridine Phosphorylase from Salmonella Typhimurium in Complex with 5-Fluorouracil at 2.15 A Resolution | | Descriptor: | 1,2-ETHANEDIOL, 5-FLUOROURACIL, GLYCEROL, ... | | Authors: | Lashkov, A.A, Sotnichenko, S.E, Prokofev, I.I, Gabdoulkhakov, A.G, Mikhailov, A.M. | | Deposit date: | 2012-03-07 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | X-ray structure of Salmonella typhimurium uridine phosphorylase complexed with 5-fluorouracil and molecular modelling of the complex of 5-fluorouracil with uridine phosphorylase from Vibrio cholerae.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
9C6P
 
 | | X-ray crystal structure of AmpC beta-lactamase with inhibitor | | Descriptor: | 3-chloro-N-(5-chloro-2-methyl-1,3-benzothiazol-6-yl)-2-hydroxybenzene-1-sulfonamide, AmpC Beta-lactamase | | Authors: | Liu, F, Shoichet, B.K. | | Deposit date: | 2024-06-08 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.663 Å) | | Cite: | Improved correlations with score, hit-rate, and affinity as docking library and testing scale increase
To Be Published
|
|
1KKX
 
 | | Solution structure of the DNA-binding domain of ADR6 | | Descriptor: | Transcription regulatory protein ADR6 | | Authors: | Tu, X, Wu, J, Xu, Y, Shi, Y. | | Deposit date: | 2001-12-10 | | Release date: | 2002-07-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | 1H, 13C and 15N resonance assignments and secondary structure of ADR6 DNA-binding domain.
J.Biomol.Nmr, 21, 2001
|
|
1BXN
 
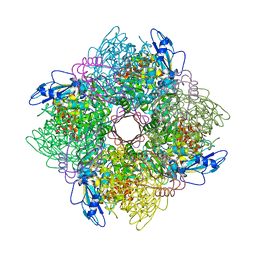 | | THE CRYSTAL STRUCTURE OF RUBISCO FROM ALCALIGENES EUTROPHUS TO 2.7 ANGSTROMS. | | Descriptor: | PHOSPHATE ION, PROTEIN (RIBULOSE BISPHOSPHATE CARBOXYLASE LARGE CHAIN), PROTEIN (RIBULOSE BISPHOSPHATE CARBOXYLASE SMALL CHAIN) | | Authors: | Hansen, S, Vollan, V.B, Hough, E, Andersen, K. | | Deposit date: | 1998-10-06 | | Release date: | 1999-10-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of rubisco from Alcaligenes eutrophus reveals a novel central eight-stranded beta-barrel formed by beta-strands from four subunits.
J.Mol.Biol., 288, 1999
|
|
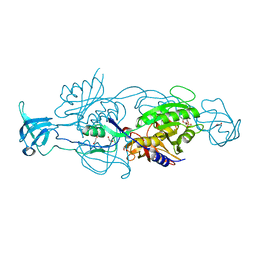
6ETF
 
 | |
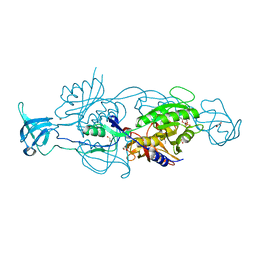
6ETH
 
 | |
6C2C
 
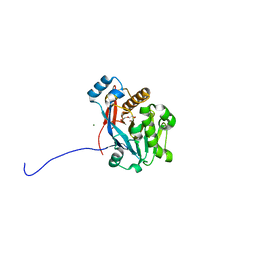 | | The molecular basis for the functional evolution of an organophosphate hydrolysing enzyme | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ZINC ION, ... | | Authors: | Hong, N.-S, Jackson, C.J, Carr, P.D, Tokuriki, N, Baier, F, Yang, G. | | Deposit date: | 2018-01-08 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.597 Å) | | Cite: | Higher-order epistasis shapes the fitness landscape of a xenobiotic-degrading enzyme.
Nat.Chem.Biol., 15, 2019
|
|
2PEL
 
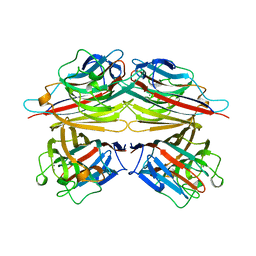 | | PEANUT LECTIN | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, PEANUT LECTIN, ... | | Authors: | Banerjee, R, Das, K, Ravishankar, R, Suguna, K, Surolia, A, Vijayan, M. | | Deposit date: | 1995-08-23 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Conformation, protein-carbohydrate interactions and a novel subunit association in the refined structure of peanut lectin-lactose complex.
J.Mol.Biol., 259, 1996
|
|
4GLJ
 
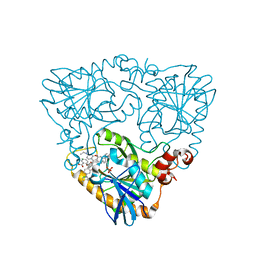 | | Crystal structure of methylthioadenosine phosphorylase in complex with rhodamine B | | Descriptor: | CHLORIDE ION, N-[9-(2-carboxyphenyl)-6-(diethylamino)-3H-xanthen-3-ylidene]-N-ethylethanaminium, PHOSPHATE ION, ... | | Authors: | Bujacz, A, Bujacz, G, Cieslinski, H, Bartasun, P. | | Deposit date: | 2012-08-14 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A study on the interaction of rhodamine B with methylthioadenosine phosphorylase protein sourced from an antarctic soil metagenomic library.
Plos One, 8, 2013
|
|
1EC2
 
 | | HIV-1 protease in complex with the inhibitor BEA428 | | Descriptor: | HIV-1 PROTEASE, N,N-[2,5-O-[DI-4-PYRIDIN-3-YL-BENZYL]-GLUCARYL]-DI-[VALYL-AMIDO-METHANE] | | Authors: | Unge, T. | | Deposit date: | 2000-01-25 | | Release date: | 2002-06-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Optimization of P1-P3 groups in symmetric and asymmetric HIV-1 protease inhibitors
Eur.J.Biochem., 270, 2003
|
|
2X7N
 
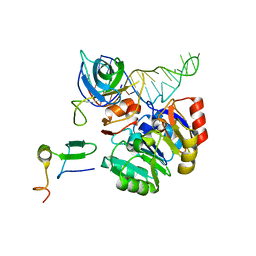 | | Mechanism of eIF6s anti-association activity | | Descriptor: | 60S RIBOSOMAL PROTEIN L23, 60S RIBOSOMAL PROTEIN L24-A, EUKARYOTIC TRANSLATION INITIATION FACTOR 6, ... | | Authors: | Gartmann, M, Blau, M, Armache, J.-P, Mielke, T, Topf, M, Beckmann, R. | | Deposit date: | 2010-03-02 | | Release date: | 2010-03-31 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (11.8 Å) | | Cite: | Mechanism of Eif6-Mediated Inhibition of Ribosomal Subunit Joining.
J.Biol.Chem., 285, 2010
|
|
1EC0
 
 | | HIV-1 protease in complex with the inhibitor bea403 | | Descriptor: | HIV-1 PROTEASE, N,N-[2,5-O-DI-2-FLUORO-BENZYL-GLUCARYL]-DI-[1-AMINO-INDAN-2-OL] | | Authors: | Unge, T. | | Deposit date: | 2000-01-25 | | Release date: | 2002-06-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Symmetric fluoro-substituted diol-based HIV protease inhibitors. Ortho-fluorinated and meta-fluorinated P1/P1'-benzyloxy side groups significantly improve the antiviral activity and preserve binding efficacy
Eur.J.Biochem., 271, 2004
|
|
3X1J
 
 | |
3X1M
 
 | |
1CGL
 
 | | Structure of the catalytic domain of fibroblast collagenase complexed with an inhibitor | | Descriptor: | CALCIUM ION, FIBROBLAST COLLAGENASE, N-[(1S)-3-{[(benzyloxy)carbonyl]amino}-1-carboxypropyl]-L-leucyl-N-(2-morpholin-4-ylethyl)-L-phenylalaninamide, ... | | Authors: | Lovejoy, B, Cleasby, A, Hassell, A.M, Longley, K, Luther, M.A, Weigl, D, Mcgeehan, G, Mcelroy, A.B, Drewry, D, Lambert, M.H, Jordan, S.R. | | Deposit date: | 1993-11-17 | | Release date: | 1995-02-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the catalytic domain of fibroblast collagenase complexed with an inhibitor.
Science, 263, 1994
|
|
4HXW
 
 | | Pyrrolopyrimidine inhibitors of dna gyrase b and topoisomerase iv, part i: structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity. | | Descriptor: | (3R)-1-[5-chloro-6-ethyl-2-(pyrido[2,3-b]pyrazin-7-ylsulfanyl)-7H-pyrrolo[2,3-d]pyrimidin-4-yl]pyrrolidin-3-amine, DNA gyrase subunit B, TERTIARY-BUTYL ALCOHOL | | Authors: | Bensen, D.C, Trzoss, M, Tari, L.W. | | Deposit date: | 2012-11-12 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Pyrrolopyrimidine inhibitors of DNA gyrase B (GyrB) and topoisomerase IV (ParE). Part I: Structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3X1K
 
 | |
1CLX
 
 | | CATALYTIC CORE OF XYLANASE A | | Descriptor: | CALCIUM ION, XYLANASE A | | Authors: | Harris, G.W, Jenkins, J.A, Connerton, I, Pickersgill, R.W. | | Deposit date: | 1995-08-31 | | Release date: | 1996-06-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Refined crystal structure of the catalytic domain of xylanase A from Pseudomonas fluorescens at 1.8 A resolution.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
4NI3
 
 | | Crystal Structure of GH29 family alpha-L-fucosidase from Fusarium graminearum in the closed form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-fucosidase GH29, ... | | Authors: | Cao, H, Walton, J.D, Brumm, P, Phillips Jr, G.N. | | Deposit date: | 2013-11-05 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.3993 Å) | | Cite: | Structure and Substrate Specificity of a Eukaryotic Fucosidase from Fusarium graminearum.
J.Biol.Chem., 289, 2014
|
|
