8T6H
 
 | |
8T83
 
 | |
6GBH
 
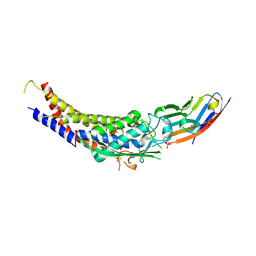 | | Helicobacter pylori adhesin HopQ type II bound to the N-terminal domain of human CEACAM1 | | Descriptor: | Carcinoembryonic antigen-related cell adhesion molecule 1, HopQ, SULFATE ION | | Authors: | Moonens, K, Kruse, T, Gerhard, M, Remaut, H. | | Deposit date: | 2018-04-13 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Helicobacter pyloriadhesin HopQ disruptstransdimerization in human CEACAMs.
EMBO J., 37, 2018
|
|
6G7X
 
 | |
6GMM
 
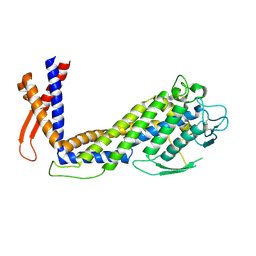 | | Crystal structure of Helicobacter pylori adhesin LabA | | Descriptor: | adhesin LabA | | Authors: | Paraskevopoulou, V, Overman, R.C, Stolnik, S, Winkler, S, Gellert, P, Falcone, F.H, Schimpl, M. | | Deposit date: | 2018-05-27 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural and binding characterization of the LacdiNAc-specific adhesin (LabA; HopD) exodomain from Helicobacter pylori
Current Research in Structural Biology, 2021
|
|
8HRV
 
 | | dutpase of helicobacter pylori 26695 | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Kumari, K, Gourinath, S. | | Deposit date: | 2022-12-16 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | dutpase of helicobacter pylori 26695
To Be Published
|
|
8II1
 
 | | Crystal structure of V30M-TTR in complex with BID | | Descriptor: | Benziodarone, Transthyretin | | Authors: | Yokoyama, T. | | Deposit date: | 2023-02-24 | | Release date: | 2023-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.907 Å) | | Cite: | Benziodarone and 6-hydroxybenziodarone are potent and selective inhibitors of transthyretin amyloidogenesis.
Bioorg.Med.Chem., 90, 2023
|
|
4EHI
 
 | | An X-ray Crystal Structure of a putative Bifunctional Phosphoribosylaminoimidazolecarboxamide Formyltransferase/IMP Cyclohydrolase | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bifunctional purine biosynthesis protein PurH, SULFATE ION | | Authors: | Brunzelle, J.S, Wawrzak, Z, Onopriyenko, O, Kwok, J, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-02 | | Release date: | 2012-06-06 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | An X-ray Crystal Structure of a putative Bifunctional Phosphoribosylaminoimidazolecarboxamide Formyltransferase/IMP Cyclohydrolase
TO BE PUBLISHED
|
|
6LNR
 
 | | Structure of intact chitinase with hevein domain from the plant Simarouba glauca, known for its traditional anti-inflammatory efficacy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(3-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | KanalElamparithi, B, Ramya, K.S, Ankur, T, Radha, A, Gunasekaran, K. | | Deposit date: | 2020-01-01 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structure of intact chitinase with hevein domain from the plant Simarouba glauca, known for its traditional anti-inflammatory efficacy.
Int.J.Biol.Macromol., 161, 2020
|
|
6IUB
 
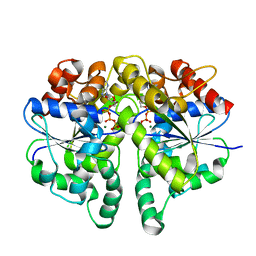 | | Structure of Helicobacter pylori Soj protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, SpoOJ regulator (Soj) | | Authors: | Chu, C.H, Yen, C.Y, Sun, Y.J. | | Deposit date: | 2018-11-28 | | Release date: | 2019-02-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Crystal structures of HpSoj-DNA complexes and the nucleoid-adaptor complex formation in chromosome segregation.
Nucleic Acids Res., 47, 2019
|
|
6IWY
 
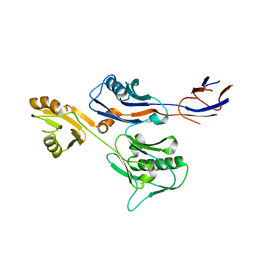 | |
6JHO
 
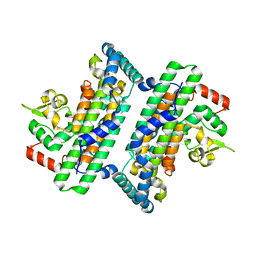 | | The complex crystal structure of Cagbeta with CagZ revealed a novel regulatory mechanism for T4SS coupling ATPase in Helicobacter pylori | | Descriptor: | Cag pathogenicity island protein (Cag5), Cag pathogenicity island protein (Cag6) | | Authors: | Wu, X, Zhao, Y, Sun, L, Ye, X, Jiang, M, Wang, Q, Wang, Q, Wu, Y. | | Deposit date: | 2019-02-18 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The complex crystal structure of Cagbeta with CagZ revealed a novel regulatory mechanism in VirD4 coupling ATPase
To Be Published
|
|
6JSX
 
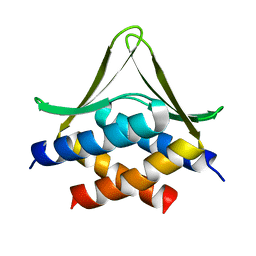 | |
6IQT
 
 | | Crystal Structure of CagV, a VirB8 homolog of T4SS from Helicobacter pylori Strain 26695 | | Descriptor: | Cag pathogenicity island protein (Cag10) | | Authors: | Wu, X, Zhao, Y, Sun, L, Jiang, M, Wang, Q, Wang, Q, Yang, W, Wu, Y. | | Deposit date: | 2018-11-08 | | Release date: | 2019-11-13 | | Last modified: | 2020-04-29 | | Method: | X-RAY DIFFRACTION (1.922 Å) | | Cite: | Crystal structure of CagV, the Helicobacter pylori homologue of the T4SS protein VirB8.
Febs J., 286, 2019
|
|
6IUD
 
 | | Structure of Helicobacter pylori Soj-ADP complex bound to DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(P*AP*GP*GP*GP*TP*GP*TP*TP*CP*CP*AP*CP*GP*TP*GP*AP*AP*AP*CP*AP*GP*GP*GP*A)-3'), DNA (5'-D(P*TP*CP*CP*CP*TP*GP*TP*TP*TP*CP*AP*CP*GP*TP*GP*GP*AP*AP*CP*AP*CP*CP*CP*T)-3'), ... | | Authors: | Yen, C.Y, Chu, C.H, Sun, Y.J. | | Deposit date: | 2018-11-28 | | Release date: | 2019-02-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.506 Å) | | Cite: | Crystal structures of HpSoj-DNA complexes and the nucleoid-adaptor complex formation in chromosome segregation.
Nucleic Acids Res., 47, 2019
|
|
6IUC
 
 | | Structure of Helicobacter pylori Soj-ATP complex bound to DNA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*AP*GP*GP*GP*TP*GP*TP*TP*CP*CP*AP*CP*GP*TP*GP*AP*AP*AP*CP*AP*GP*GP*GP*A)-3'), DNA (5'-D(P*TP*CP*CP*CP*TP*GP*TP*TP*TP*CP*AP*CP*GP*TP*GP*GP*AP*AP*CP*AP*CP*CP*CP*T)-3'), ... | | Authors: | Chu, C.H, Yen, C.Y, Sun, Y.J. | | Deposit date: | 2018-11-28 | | Release date: | 2019-02-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structures of HpSoj-DNA complexes and the nucleoid-adaptor complex formation in chromosome segregation.
Nucleic Acids Res., 47, 2019
|
|
6IHC
 
 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase(FabZ) Y100A mutant in complex with holo-ACP from Helicobacter pylori | | Descriptor: | 3-hydroxyacyl-[acyl-carrier-protein] dehydratase FabZ, CITRIC ACID, N~3~-[(2S)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-N-(2-sulfanylethyl)-beta-alaninamide, ... | | Authors: | Shen, S.Q, Zhang, L, Zhang, L. | | Deposit date: | 2018-09-29 | | Release date: | 2019-04-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A back-door Phenylalanine coordinates the stepwise hexameric loading of acyl carrier protein by the fatty acid biosynthesis enzyme beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ).
Int. J. Biol. Macromol., 128, 2019
|
|
5NBD
 
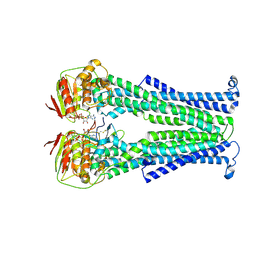 | | PglK flippase in complex with inhibitory nanobody | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Nanobody, WlaB protein | | Authors: | Perez, C, Pardon, E, Steyaert, J, Locher, K.P. | | Deposit date: | 2017-03-01 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structural basis of inhibition of lipid-linked oligosaccharide flippase PglK by a conformational nanobody.
Sci Rep, 7, 2017
|
|
4DIF
 
 | | Structure of A1-type ketoreductase | | Descriptor: | AmphB, GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zheng, J, Keatinge-Clay, A.T. | | Deposit date: | 2012-01-30 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.521 Å) | | Cite: | Structure and mutagenesis of A2-type ketoreductase from modular polyketide synthase reveals insights into stereospecificity
To be Published
|
|
5MJI
 
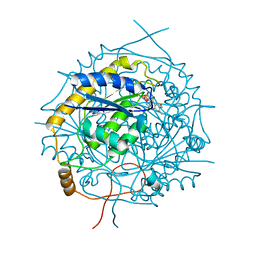 | | Crystal Structure of RosB with bound intermediate OHC-RP (8-demethyl-8-formylriboflavin-5'-phosphate) | | Descriptor: | BRAMP domain protein, [(2~{S},3~{R},4~{R})-5-[8-methanoyl-7-methyl-2,4-bis(oxidanylidene)-1~{H}-benzo[g]pteridin-10-ium-10-yl]-2,3,4-tris(oxidanyl)pentyl] dihydrogen phosphate | | Authors: | Konjik, V, Bruenle, S, Demmer, U, Vanselow, A, Sandhoff, R, Mack, M, Ermler, U. | | Deposit date: | 2016-12-01 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of RosB: Insights into the Reaction Mechanism of the First Member of a Family of Flavodoxin-like Enzymes.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
4DWT
 
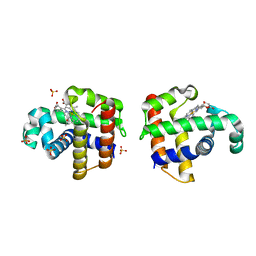 | |
4DWU
 
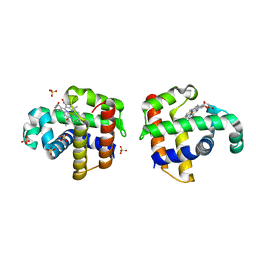 | |
7E69
 
 | | The crystal structure of peptidoglycan peptidase in complex with inhibitor 3-3 | | Descriptor: | N-oxidanyl-4-[(4-sulfamoylphenyl)methyl]benzamide, Peptidase M23, ZINC ION | | Authors: | Choi, Y, Min, K.J, Yoon, H.J, Lee, H.H. | | Deposit date: | 2021-02-21 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structure-based inhibitor design for reshaping bacterial morphology
Commun Biol, 5, 2022
|
|
7E65
 
 | | The crystal structure of peptidoglycan peptidase in complex with inhibitor 3 | | Descriptor: | (2S)-2-acetamido-N-[(2R)-1-(oxidanylamino)-1-oxidanylidene-propan-2-yl]-3-(4-sulfamoylphenyl)propanamide, Peptidase M23, ZINC ION | | Authors: | Choi, Y, Min, K.J, Yoon, H.J, Lee, H.H. | | Deposit date: | 2021-02-21 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure-based inhibitor design for reshaping bacterial morphology
Commun Biol, 5, 2022
|
|
7E60
 
 | | The crystal structure of peptidoglycan peptidase in complex with inhibitor 1 | | Descriptor: | (2~{R},6~{S})-2,6-diacetamido-7-[[(2~{R})-1-(oxidanylamino)-1-oxidanylidene-propan-2-yl]amino]-7-oxidanylidene-heptanoic acid, Peptidase M23, ZINC ION | | Authors: | Min, K, Yoon, H.J, Choi, Y, Lee, H.H. | | Deposit date: | 2021-02-21 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structure-based inhibitor design for reshaping bacterial morphology
Commun Biol, 5, 2022
|
|
