8T8T
 
 | |
4OB0
 
 | | Crystal Structure of Nitrile Hydratase from Pseudonocardia thermophila bound to Phenyl Boronic Acid | | Descriptor: | COBALT (II) ION, Cobalt-containing nitrile hydratase subunit alpha, Cobalt-containing nitrile hydratase subunit beta, ... | | Authors: | Rui, W, Salette, M, Ruslan, S, Richard, H, Dali, L. | | Deposit date: | 2014-01-06 | | Release date: | 2014-11-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The active site sulfenic acid ligand in nitrile hydratases can function as a nucleophile.
J.Am.Chem.Soc., 136, 2014
|
|
5CDW
 
 | | Crystal Structure Analysis of a mutant Grb2 SH2 domain (W121G) with a pYVNV peptide | | Descriptor: | Growth factor receptor-bound protein 2, SER-PTR-VAL-ASN-VAL-GLN | | Authors: | Papaioannou, D, Geibel, S, Kunze, M, Kay, C, Waksman, G. | | Deposit date: | 2015-07-05 | | Release date: | 2016-05-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | Structural and biophysical investigation of the interaction of a mutant Grb2 SH2 domain (W121G) with its cognate phosphopeptide.
Protein Sci., 25, 2016
|
|
3FO7
 
 | | Simultaneous inhibition of anti-coagulation and inflammation: Crystal structure of phospholipase A2 complexed with indomethacin at 1.4 A resolution reveals the presence of the new common ligand binding site | | Descriptor: | INDOMETHACIN, Phospholipase A2 VRV-PL-VIIIa, SULFATE ION | | Authors: | Singh, N, Prem Kumar, R, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2008-12-29 | | Release date: | 2009-01-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Simultaneous inhibition of anti-coagulation and inflammation: Crystal structure of phospholipase A2 complexed with indomethacin at 1.4 A resolution reveals the presence of the new common ligand binding site
To be Published
|
|
4YGO
 
 | | Dodecameric structure of spermidine N-acetyltransferase from Vibrio cholerae in intermediate state | | Descriptor: | CALCIUM ION, METHANOL, Spermidine n1-acetyltransferase | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-02-26 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Substrate-Induced Allosteric Change in the Quaternary Structure of the Spermidine N-Acetyltransferase SpeG.
J.Mol.Biol., 427, 2015
|
|
2ETZ
 
 | | The NMR minimized average structure of the Itk SH2 domain bound to a phosphopeptide | | Descriptor: | Lymphocyte cytosolic protein 2 phosphopeptide fragment, Tyrosine-protein kinase ITK/TSK | | Authors: | Sundd, M, Pletneva, E.V, Fulton, D.B, Andreotti, A.H. | | Deposit date: | 2005-10-27 | | Release date: | 2006-02-07 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Molecular Details of Itk Activation by Prolyl Isomerization and Phospholigand Binding: The NMR Structure of the Itk SH2 Domain Bound to a Phosphopeptide.
J.Mol.Biol., 357, 2006
|
|
5HRW
 
 | | Crystal structure of the fifth bromodomain of human PB1 in complex with 1-propylisochromeno[3,4-c]pyrazol-5(2H)-one) compound | | Descriptor: | 1,2-ETHANEDIOL, 1-propylisochromeno[3,4-c]pyrazol-5(3H)-one, Protein polybromo-1, ... | | Authors: | Tallant, C, Myrianthopoulos, V, Gaboriaud-Kolar, N, Newman, J.A, Picaud, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Mikros, E, Knapp, S. | | Deposit date: | 2016-01-24 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery and Optimization of a Selective Ligand for the Switch/Sucrose Nonfermenting-Related Bromodomains of Polybromo Protein-1 by the Use of Virtual Screening and Hydration Analysis.
J.Med.Chem., 59, 2016
|
|
5FTA
 
 | | Crystal structure of the N-terminal BTB domain of human KCTD10 | | Descriptor: | BTB/POZ DOMAIN-CONTAINING ADAPTER FOR CUL3-MEDIATED RHOA DEGRADATION PROTEIN 3, MERCURY (II) ION | | Authors: | Pinkas, D.M, Sanvitale, C.E, Solcan, N, Goubin, S, Tallant, C, Newman, J.A, Kopec, J, Fitzpatrick, F, Talon, R, Collins, P, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2016-01-12 | | Release date: | 2016-02-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structural complexity in the KCTD family of Cullin3-dependent E3 ubiquitin ligases.
Biochem. J., 474, 2017
|
|
5HVO
 
 | | Structure of Aspergillus fumigatus trehalose-6-phosphate synthase B in complex with UDP and validoxylamine A | | Descriptor: | (1S,2S,3R,6S)-4-(HYDROXYMETHYL)-6-{[(1S,2S,3S,4R,5R)-2,3,4-TRIHYDROXY-5-(HYDROXYMETHYL)CYCLOHEXYL]AMINO}CYCLOHEX-4-ENE-1,2,3-TRIOL, Alpha,alpha-trehalose-phosphate synthase (UDP-forming), URIDINE-5'-DIPHOSPHATE | | Authors: | Miao, Y, Brennan, R.G. | | Deposit date: | 2016-01-28 | | Release date: | 2017-05-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.467 Å) | | Cite: | Structural and In Vivo Studies on Trehalose-6-Phosphate Synthase from Pathogenic Fungi Provide Insights into Its Catalytic Mechanism, Biological Necessity, and Potential for Novel Antifungal Drug Design.
MBio, 8, 2017
|
|
4RPH
 
 | |
5N5W
 
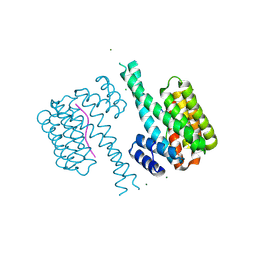 | | 14-3-3 sigma in complex with TAZ pS89 peptide and fragment NV3 | | Descriptor: | 14-3-3 protein sigma, 4-[3,5-bis(chloranyl)pyridin-2-yl]oxyaniline, CALCIUM ION, ... | | Authors: | Sijbesma, E, Leysen, S, Ottmann, C. | | Deposit date: | 2017-02-14 | | Release date: | 2017-07-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Identification of Two Secondary Ligand Binding Sites in 14-3-3 Proteins Using Fragment Screening.
Biochemistry, 56, 2017
|
|
5TIS
 
 | | Room temperature XFEL structure of the native, doubly-illuminated photosystem II complex | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Young, I.D, Ibrahim, M, Chatterjee, R, Gul, S, Fuller, F, Koroidov, S, Brewster, A.S, Tran, R, Alonso-Mori, R, Kroll, T, Michels-Clark, T, Laksmono, H, Sierra, R.G, Stan, C.A, Hussein, R, Zhang, M, Douthit, L, Kubin, M, de Lichtenberg, C, Pham, L.V, Nilsson, H, Cheah, M.H, Shevela, D, Saracini, C, Bean, M.A, Seuffert, I, Sokaras, D, Weng, T.-C, Pastor, E, Weninger, C, Fransson, T, Lassalle, L, Braeuer, P, Aller, P, Docker, P.T, Andi, B, Orville, A.M, Glownia, J.M, Nelson, S, Sikorski, M, Zhu, D, Hunter, M.S, Aquila, A, Koglin, J.E, Robinson, J, Liang, M, Boutet, S, Lyubimov, A.Y, Uervirojnangkoorn, M, Moriarty, N.W, Liebschner, D, Afonine, P.V, Watermann, D.G, Evans, G, Wernet, P, Dobbek, H, Weis, W.I, Brunger, A.T, Zwart, P.H, Adams, P.D, Zouni, A, Messinger, J, Bergmann, U, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2016-10-03 | | Release date: | 2016-11-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25000381 Å) | | Cite: | Structure of photosystem II and substrate binding at room temperature.
Nature, 540, 2016
|
|
4PUJ
 
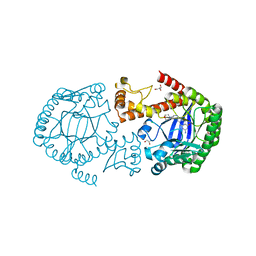 | | tRNA-Guanine Transglycosylase (TGT) in Complex with 6-Amino-2-{[2-(morpholin-4-yl)ethyl]amino}-1H,7H,8H-imidazo[4,5-g]quinazolin-8-one | | Descriptor: | 6-amino-2-[(2-morpholin-4-ylethyl)amino]-3,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one, CHLORIDE ION, GLYCEROL, ... | | Authors: | Neeb, M, Heine, A, Klebe, G. | | Deposit date: | 2014-03-13 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Chasing Protons: How Isothermal Titration Calorimetry, Mutagenesis, and pKa Calculations Trace the Locus of Charge in Ligand Binding to a tRNA-Binding Enzyme.
J.Med.Chem., 57, 2014
|
|
1JQQ
 
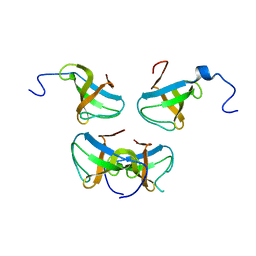 | | Crystal structure of Pex13p(301-386) SH3 domain | | Descriptor: | PEROXISOMAL MEMBRANE PROTEIN PAS20 | | Authors: | Douangamath, A, Mayans, O, Barnett, P, Distel, B, Wilmanns, M. | | Deposit date: | 2001-08-08 | | Release date: | 2002-12-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Topography for Independent Binding of alpha-Helical and PPII-Helical Ligands to a Peroxisomal SH3 Domain
Mol.Cell, 10, 2002
|
|
5B5F
 
 | | Crystal structure of ALiS3-Streptavidin complex | | Descriptor: | N-methyl-3-(4-oxo-4,5-dihydrofuro[3,2-c]pyridin-2-yl)benzenesulfonamide, Streptavidin | | Authors: | Sugiyama, S, Terai, T, Kakinouchi, K, Fujikake, R, Nagano, T, Urano, Y. | | Deposit date: | 2016-05-04 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Improving the Solubility of Artificial Ligands of Streptavidin to Enable More Practical Reversible Switching of Protein Localization in Cells
Chembiochem, 18, 2017
|
|
4LP2
 
 | |
1Z2K
 
 | | NMR structure of the D1 domain of the Natural Killer Cell Receptor, 2B4 | | Descriptor: | Natural killer cell receptor 2B4 | | Authors: | Ames, J.B, Vyas, V, Lusin, J.D, Mariuzza, R. | | Deposit date: | 2005-03-08 | | Release date: | 2005-05-03 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Natural Killer Cell Receptor 2B4:
Implications for Ligand Recognition
Biochemistry, 44, 2005
|
|
5MH2
 
 | |
1Z8Q
 
 | | Ferrous dioxygen complex of the A245T cytochrome P450eryF | | Descriptor: | 6-DEOXYERYTHRONOLIDE B, 6-deoxyerythronolide B hydroxylase, OXYGEN MOLECULE, ... | | Authors: | Nagano, S, Cupp-Vickery, J.R, Poulos, T.L. | | Deposit date: | 2005-03-31 | | Release date: | 2005-04-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the ferrous dioxygen complex of wild-type cytochrome P450eryF and its mutants, A245S and A245T: investigation of the proton transfer system in P450eryF.
J.Biol.Chem., 280, 2005
|
|
1DLG
 
 | | CRYSTAL STRUCTURE OF THE C115S ENTEROBACTER CLOACAE MURA IN THE UN-LIGANDED STATE | | Descriptor: | CYCLOHEXYLAMMONIUM ION, PHOSPHATE ION, UDP-N-ACETYLGLUCOSAMINE ENOLPYRUVYL TRANSFERASE MURA | | Authors: | Schonbrunn, E, Eschenburg, S, Krekel, F, Luger, K, Amrhein, N. | | Deposit date: | 1999-12-09 | | Release date: | 2000-04-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Role of the loop containing residue 115 in the induced-fit mechanism of the bacterial cell wall biosynthetic enzyme MurA.
Biochemistry, 39, 2000
|
|
5MH0
 
 | |
1Z8O
 
 | | Ferrous dioxygen complex of the wild-type cytochrome P450eryF | | Descriptor: | 6-DEOXYERYTHRONOLIDE B, 6-deoxyerythronolide B hydroxylase, OXYGEN MOLECULE, ... | | Authors: | Nagano, S, Cupp-Vickery, J.R, Poulos, T.L. | | Deposit date: | 2005-03-31 | | Release date: | 2005-04-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of the ferrous dioxygen complex of wild-type cytochrome P450eryF and its mutants, A245S and A245T: investigation of the proton transfer system in P450eryF.
J.Biol.Chem., 280, 2005
|
|
6A0U
 
 | | Homoserine dehydrogenase K195A mutant from Thermus thermophilus HB8 complexed with HSE and NADP+ | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, FORMIC ACID, GLYCEROL, ... | | Authors: | Akai, S, Ikushiro, H, Sawai, T, Yano, T, Kamiya, N, Miyahara, I. | | Deposit date: | 2018-06-06 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | The crystal structure of homoserine dehydrogenase complexed with l-homoserine and NADPH in a closed form
J. Biochem., 165, 2019
|
|
4OB3
 
 | | Crystal Structure of Nitrile Hydratase from Pseudonocardia thermophila : A Reference Structure to Boronic Acid Inhibition of Nitrile Hydratase | | Descriptor: | COBALT (II) ION, Cobalt-containing nitrile hydratase subunit alpha, Cobalt-containing nitrile hydratase subunit beta, ... | | Authors: | Rui, W, Salette, M, Ruslan, S, Richard, H, Dali, L. | | Deposit date: | 2014-01-06 | | Release date: | 2014-11-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The active site sulfenic acid ligand in nitrile hydratases can function as a nucleophile.
J.Am.Chem.Soc., 136, 2014
|
|
4PUM
 
 | | tRNA-Guanine Transglycosylase (TGT) Mutant D156N in Complex with 6-Amino-2-(methylamino)-1H,7H,8H-imidazo[4,5-g]quinazolin-8-one | | Descriptor: | 6-amino-2-(methylamino)-3,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Neeb, M, Heine, A, Klebe, G. | | Deposit date: | 2014-03-13 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Chasing Protons: How Isothermal Titration Calorimetry, Mutagenesis, and pKa Calculations Trace the Locus of Charge in Ligand Binding to a tRNA-Binding Enzyme.
J.Med.Chem., 57, 2014
|
|
