1QM8
 
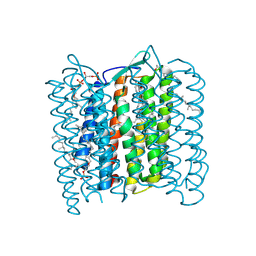 | | Structure of Bacteriorhodopsin at 100 K | | Descriptor: | 2,3-DI-O-PHYTANLY-3-SN-GLYCERO-1-PHOSPHORYL-3'-SN-GLYCEROL-1'-PHOSPHATE, 2,3-DI-PHYTANYL-GLYCEROL, 3-PHOSPHORYL-[1,2-DI-PHYTANYL]GLYCEROL, ... | | Authors: | Takeda, K, Matsui, Y, Sato, H, Hino, T, Kanamori, E, Okumura, H, Yamane, T, Kamiya, N, Kouyama, T. | | Deposit date: | 1999-09-22 | | Release date: | 2000-08-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Novel Three-Dimensional Crystal of Bacteriorhodopsin Obtained by Successive Fusion of the Vesicular Assemblies.
J.Mol.Biol., 283, 1998
|
|
1F59
 
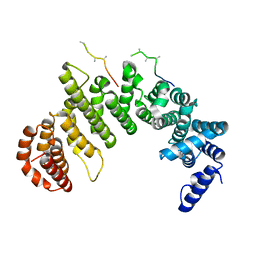 | | IMPORTIN-BETA-FXFG NUCLEOPORIN COMPLEX | | Descriptor: | FXFG NUCLEOPORIN REPEATS, IMPORTIN BETA-1 | | Authors: | Bayliss, R, Littlewood, T, Stewart, M. | | Deposit date: | 2000-06-13 | | Release date: | 2000-08-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the interaction between FxFG nucleoporin repeats and importin-beta in nuclear trafficking.
Cell(Cambridge,Mass.), 102, 2000
|
|
1QKT
 
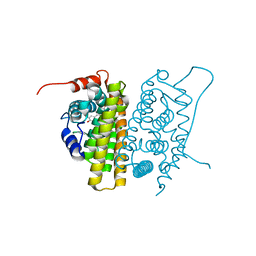 | | MUTANT ESTROGEN NUCLEAR RECEPTOR LIGAND BINDING DOMAIN COMPLEXED WITH ESTRADIOL | | Descriptor: | ESTRADIOL, ESTRADIOL RECEPTOR | | Authors: | Ruff, M, Gangloff, M, Eiler, S, Duclaud, S, Wurtz, J.M, Moras, D. | | Deposit date: | 1999-08-05 | | Release date: | 2000-08-18 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of a Mutant Heralpha Ligand- Binding Domain Reveals Key Structural Features for the Mechanism of Partial Agonism
J.Biol.Chem., 276, 2001
|
|
1QKU
 
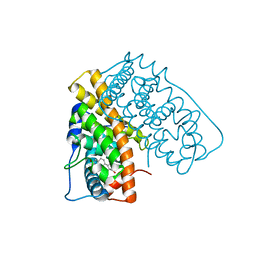 | | WILD TYPE ESTROGEN NUCLEAR RECEPTOR LIGAND BINDING DOMAIN COMPLEXED WITH ESTRADIOL | | Descriptor: | ESTRADIOL, ESTRADIOL RECEPTOR | | Authors: | Ruff, M, Gangloff, M, Eiler, S, Duclaud, S, Wurtz, J.M, Moras, D. | | Deposit date: | 1999-08-05 | | Release date: | 2000-08-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of a mutant hERalpha ligand-binding domain reveals key structural features for the mechanism of partial agonism.
J. Biol. Chem., 276, 2001
|
|
1DY3
 
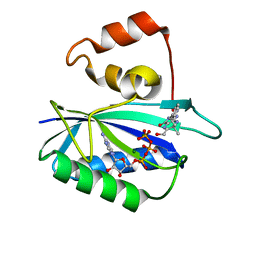 | | Ternary complex of 7,8-dihydro-6-hydroxymethylpterinpyrophosphokinase from Escherichia coli with ATP and a substrate analogue. | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 7,8-DIHYDRO-6-HYDROXYMETHYL-7-METHYL-7-[2-PHENYLETHYL]-PTERIN, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Stammers, D.K, Achari, A, Somers, D.O, Bryant, P.K, Rosemond, J, Scott, D.L, Champness, J.N. | | Deposit date: | 2000-01-21 | | Release date: | 2000-08-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.0A X-Ray Structure of the Ternary Complex of 7,8-Dihydro-6-Hydroxymethylpterinpyrophosphokinase from Escherichia Coli with ATP and a Substrate Analogue
FEBS Lett., 456, 1999
|
|
1E15
 
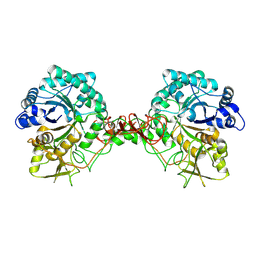 | | Chitinase B from Serratia Marcescens | | Descriptor: | CHITINASE B | | Authors: | Van Aalten, D.M.F, Synstad, B, Brurberg, M.B, Hough, E, Riise, B.W, Eijsink, V.G.H, Wierenga, R.K. | | Deposit date: | 2000-04-18 | | Release date: | 2000-08-18 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a Two-Domain Chitotriosidase from Serratia Marcescens at 1.9 Angstrom Resoltuion
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1DYL
 
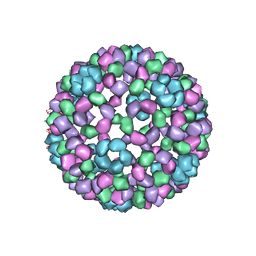 | | 9 ANGSTROM RESOLUTION CRYO-EM RECONSTRUCTION STRUCTURE OF SEMLIKI FOREST VIRUS (SFV) AND FITTING OF THE CAPSID PROTEIN STRUCTURE IN THE EM DENSITY | | Descriptor: | NUCLEOCAPSID PROTEIN | | Authors: | Mancini, E.J, Clarke, M, Gowen, B.E, Rutten, T, Fuller, S.D. | | Deposit date: | 2000-02-02 | | Release date: | 2000-08-18 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Cryo-Electron Microscopy Reveals the Functional Organization of an Enveloped Virus, Semliki Forest Virus.
Mol.Cell, 5, 2000
|
|
1QKW
 
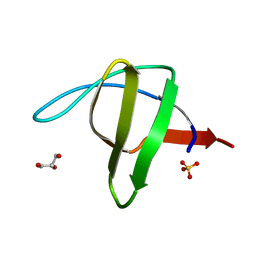 | | Alpha-spectrin Src Homology 3 domain, N47G mutant in the distal loop. | | Descriptor: | ALPHA II SPECTRIN, GLYCEROL, SULFATE ION | | Authors: | Vega, M.C, Martinez, J, Serrano, L. | | Deposit date: | 1999-08-16 | | Release date: | 2000-08-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Thermodynamic and structural characterization of Asn and Ala residues in the disallowed II' region of the Ramachandran plot.
Protein Sci., 9, 2000
|
|
1E0R
 
 | | Beta-apical domain of thermosome | | Descriptor: | THERMOSOME | | Authors: | Bosch, G, Baumeister, W, Essen, L.-O. | | Deposit date: | 2000-04-06 | | Release date: | 2000-08-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Beta-Apical Domain from Thermosome Reveals Structural Plasticity in Protrusion Region
J.Mol.Biol., 301, 2000
|
|
1E0P
 
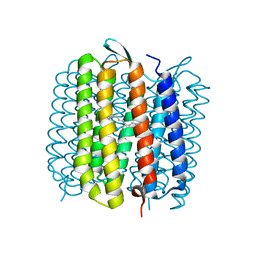 | | L intermediate of bacteriorhodopsin | | Descriptor: | BACTERIORHODOPSIN, GROUND STATE, RETINAL | | Authors: | Royant, A, Edman, K, Ursby, T, Pebay-Peyroula, E, Landau, E.M, Neutze, R. | | Deposit date: | 2000-04-04 | | Release date: | 2000-08-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Helix Deformation is Coupled to Vectorial Proton Transport in Bacteriorhodopsin'S Photocycle
Nature, 406, 2000
|
|
1EM0
 
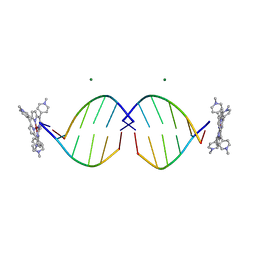 | | COMPLEX OF D(CCTAGG) WITH TETRA-[N-METHYL-PYRIDYL] PORPHYRIN | | Descriptor: | DNA (5'-D(*(CBR)P*CP*TP*AP*GP*G)-3'), MAGNESIUM ION, TETRA[N-METHYL-PYRIDYL] PORPHYRIN-NICKEL | | Authors: | Neidle, S, Sanderson, M, Bennett, M, Krah, A, Wien, F, Garman, E, McKenna, R. | | Deposit date: | 2000-03-14 | | Release date: | 2000-08-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | A DNA-porphyrin minor-groove complex at atomic resolution: the structural consequences of porphyrin ruffling.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1FF2
 
 | |
1E42
 
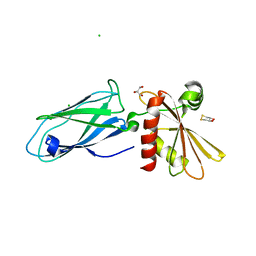 | | Beta2-adaptin appendage domain, from clathrin adaptor AP2 | | Descriptor: | AP-2 COMPLEX SUBUNIT BETA, CHLORIDE ION, DITHIANE DIOL, ... | | Authors: | Owen, D.J, Evans, P.R, McMahon, H.T. | | Deposit date: | 2000-06-27 | | Release date: | 2000-08-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structure and Function of the Beta2-Adaptin Appendage Domain
Embo J., 19, 2000
|
|
1F5U
 
 | |
1EG3
 
 | | STRUCTURE OF A DYSTROPHIN WW DOMAIN FRAGMENT IN COMPLEX WITH A BETA-DYSTROGLYCAN PEPTIDE | | Descriptor: | DYSTROPHIN | | Authors: | Huang, X, Poy, F, Zhang, R, Joachimiak, A, Sudol, M, Eck, M.J. | | Deposit date: | 2000-02-11 | | Release date: | 2000-08-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a WW domain containing fragment of dystrophin in complex with beta-dystroglycan.
Nat.Struct.Biol., 7, 2000
|
|
1EG4
 
 | | STRUCTURE OF A DYSTROPHIN WW DOMAIN FRAGMENT IN COMPLEX WITH A BETA-DYSTROGLYCAN PEPTIDE | | Descriptor: | BETA-DYSTROGLYCAN, DYSTROPHIN | | Authors: | Huang, X, Poy, F, Zhang, R, Joachimiak, A, Sudol, M, Eck, M.J. | | Deposit date: | 2000-02-11 | | Release date: | 2000-08-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a WW domain containing fragment of dystrophin in complex with beta-dystroglycan.
Nat.Struct.Biol., 7, 2000
|
|
1F0X
 
 | | CRYSTAL STRUCTURE OF D-LACTATE DEHYDROGENASE, A PERIPHERAL MEMBRANE RESPIRATORY ENZYME. | | Descriptor: | D-LACTATE DEHYDROGENASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Dym, O, Pratt, E.A, Ho, C, Eisenberg, D. | | Deposit date: | 2000-05-17 | | Release date: | 2000-08-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of D-lactate dehydrogenase, a peripheral membrane respiratory enzyme.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1F51
 
 | | A TRANSIENT INTERACTION BETWEEN TWO PHOSPHORELAY PROTEINS TRAPPED IN A CRYSTAL LATTICE REVEALS THE MECHANISM OF MOLECULAR RECOGNITION AND PHOSPHOTRANSFER IN SINGAL TRANSDUCTION | | Descriptor: | MAGNESIUM ION, SPORULATION INITIATION PHOSPHOTRANSFERASE B, SPORULATION INITIATION PHOSPHOTRANSFERASE F | | Authors: | Zapf, J, Sen, U, Madhusudan, M, Hoch, J.A, Varughese, K.I. | | Deposit date: | 2000-06-11 | | Release date: | 2000-08-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A transient interaction between two phosphorelay proteins trapped in a crystal lattice reveals the mechanism of molecular recognition and phosphotransfer in signal transduction.
Structure Fold.Des., 8, 2000
|
|
1FI9
 
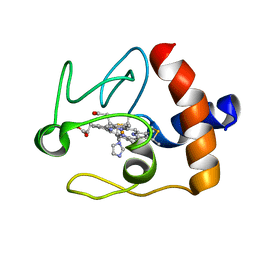 | | SOLUTION STRUCTURE OF THE IMIDAZOLE COMPLEX OF CYTOCHROME C | | Descriptor: | CYTOCHROME C, HEME C, IMIDAZOLE | | Authors: | Banci, L, Bertini, I, Liu, G, Lu, J, Reddig, T, Tang, W, Wu, Y, Zhu, D. | | Deposit date: | 2000-08-03 | | Release date: | 2000-08-23 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Effects of extrinsic imidazole ligation on the molecular and electronic structure of cytochrome c
J.Biol.Inorg.Chem., 6, 2001
|
|
1FF5
 
 | | STRUCTURE OF E-CADHERIN DOUBLE DOMAIN | | Descriptor: | CALCIUM ION, EPITHELIAL CADHERIN | | Authors: | Pertz, O, Bozic, D, Koch, A.W, Fauser, C, Brancaccio, A, Engel, J. | | Deposit date: | 2000-07-25 | | Release date: | 2000-08-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | A new crystal structure, Ca2+ dependence and mutational analysis reveal molecular details of E-cadherin homoassociation.
EMBO J., 18, 1999
|
|
1CUO
 
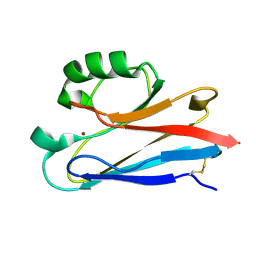 | | CRYSTAL STRUCTURE ANALYSIS OF ISOMER-2 AZURIN FROM METHYLOMONAS J | | Descriptor: | COPPER (II) ION, PROTEIN (AZURIN ISO-2) | | Authors: | Inoue, T, Nishio, N, Kai, Y, Suzuki, S, Kataoka, K. | | Deposit date: | 1999-08-21 | | Release date: | 2000-08-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The significance of the flexible loop in the azurin (Az-iso2) from the obligate methylotroph Methylomonas sp. strain J.
J.Mol.Biol., 333, 2003
|
|
1CU1
 
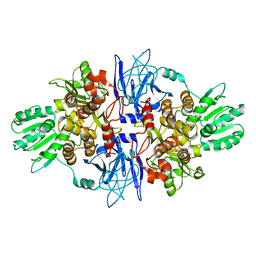 | | CRYSTAL STRUCTURE OF AN ENZYME COMPLEX FROM HEPATITIS C VIRUS | | Descriptor: | PHOSPHATE ION, PROTEIN (PROTEASE/HELICASE NS3), ZINC ION | | Authors: | Yao, N, Weber, P.C. | | Deposit date: | 1999-08-20 | | Release date: | 2000-08-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular views of viral polyprotein processing revealed by the crystal structure of the hepatitis C virus bifunctional protease-helicase.
Structure Fold.Des., 7, 1999
|
|
1FI7
 
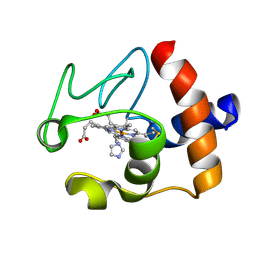 | | Solution structure of the imidazole complex of cytochrome C | | Descriptor: | CYTOCHROME C, HEME C, IMIDAZOLE | | Authors: | Banci, L, Bertini, I, Liu, G, Lu, J, Reddig, T, Tang, W, Wu, Y, Zhu, D. | | Deposit date: | 2000-08-03 | | Release date: | 2000-08-23 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Effects of extrinsic imidazole ligation on the molecular and electronic structure of cytochrome c
J.Biol.Inorg.Chem., 6, 2001
|
|
1FJ4
 
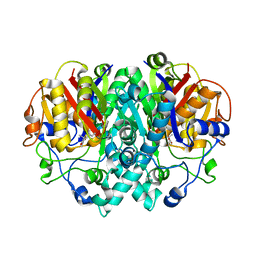 | | THE STRUCTURE OF BETA-KETOACYL-[ACYL CARRIER PROTEIN] SYNTHASE I IN COMPLEX WITH THIOLACTOMYCIN, IMPLICATIONS FOR DRUG DESIGN | | Descriptor: | BETA-KETOACYL-[ACYL CARRIER PROTEIN] SYNTHASE I, THIOLACTOMYCIN | | Authors: | Price, A.C, Choi, K, Heath, R.J, Li, Z, White, S.W, Rock, C.O. | | Deposit date: | 2000-08-07 | | Release date: | 2000-08-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Inhibition of beta-ketoacyl-acyl carrier protein synthases by thiolactomycin and cerulenin. Structure and mechanism.
J.Biol.Chem., 276, 2001
|
|
1FHG
 
 | | HIGH RESOLUTION REFINEMENT OF TELOKIN | | Descriptor: | TELOKIN | | Authors: | Tomchick, D.R, Minor, W, Kiyatkin, A, Lewinski, K, Somlyo, A.V, Somlyo, A.P. | | Deposit date: | 2000-08-01 | | Release date: | 2000-08-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure determination of telokin, the C-terminal domain of myosin light chain kinase, at 2.8 A resolution.
J.Mol.Biol., 227, 1992
|
|
