5DOZ
 
 | | Crystal structure of JamJ enoyl reductase (NADPH bound) | | Descriptor: | ACETATE ION, JamJ, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Khare, D, Smith, J.L. | | Deposit date: | 2015-09-11 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural Basis for Cyclopropanation by a Unique Enoyl-Acyl Carrier Protein Reductase.
Structure, 23, 2015
|
|
7R5D
 
 | | PARP15 catalytic domain in complex with OUL234 | | Descriptor: | 6-propan-2-yl-[1,2,4]triazolo[3,4-b][1,3]benzothiazole, DIMETHYL SULFOXIDE, Protein mono-ADP-ribosyltransferase PARP15 | | Authors: | Maksimainen, M.M, Lehtio, L. | | Deposit date: | 2022-02-10 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | [1,2,4]Triazolo[3,4- b ]benzothiazole Scaffold as Versatile Nicotinamide Mimic Allowing Nanomolar Inhibition of Different PARP Enzymes.
J.Med.Chem., 66, 2023
|
|
4K9G
 
 | | 1.55 A Crystal Structure of Macrophage Migration Inhibitory Factor bound to ISO-66 and a related compound | | Descriptor: | (4R,6Z)-6-(3-fluoro-4-hydroxyphenyl)-4-hydroxy-6-iminohexan-2-one, 1-[(5S)-3-(3-fluoro-4-hydroxyphenyl)-4,5-dihydro-1,2-oxazol-5-yl]propan-2-one, CHLORIDE ION, ... | | Authors: | Crichlow, G.V, Al-Abed, Y, Lolis, E.J. | | Deposit date: | 2013-04-19 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | ISO-66, a novel inhibitor of macrophage migration, shows efficacy in melanoma and colon cancer models.
INT J ONCOL., 45, 2014
|
|
4K5D
 
 | | Structure of neuronal nitric oxide synthase heme domain in complex with (S)-1,2-bis((2-amino-4-methylpyridin-6-yl)-methoxy)-propan-3-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6,6'-{[(2S)-3-aminopropane-1,2-diyl]bis(oxymethanediyl)}bis(4-methylpyridin-2-amine), ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2013-04-14 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Chiral linkers to improve selectivity of double-headed neuronal nitric oxide synthase inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
2KV6
 
 | | Tetrapeptide KWKK conjugated to oligonucleotide duplex by a trimethylene tether | | Descriptor: | 5'-D(*GP*CP*TP*AP*GP*CP*GP*AP*GP*TP*CP*C)-3', 5'-D(*GP*GP*AP*CP*TP*CP*GP*CP*TP*AP*GP*C)-3', KWKK Tetrapeptide | | Authors: | Huang, H, Kozekov, I.D, Kozekova, A, Rizzo, C.J, McCullough, A, LLoyd, R.S, Stone, M.P. | | Deposit date: | 2010-03-08 | | Release date: | 2010-07-21 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Minor Groove Orientation of the KWKK Peptide Tethered via the N-Terminal Amine to the Acrolein-Derived 1,N(2)-gamma-Hydroxypropanodeoxyguanosine Lesion with a Trimethylene Linkage .
Biochemistry, 49, 2010
|
|
4K5E
 
 | | Structure of neuronal nitric oxide synthase heme domain in complex with (R)-1,2-bis((2-amino-4-methylpyridin-6-yl)-methoxy)-propan-3-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6,6'-{[(2R)-3-aminopropane-1,2-diyl]bis(oxymethanediyl)}bis(4-methylpyridin-2-amine), ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2013-04-14 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Chiral linkers to improve selectivity of double-headed neuronal nitric oxide synthase inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4H81
 
 | | Crystal structure of branched-chain alpha-ketoacid dehydrogenase kinase/(R)-2-chloro-3-phenylpropanoic acid complex with ADP | | Descriptor: | (2R)-2-chloro-3-phenylpropanoic acid, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Tso, S.C, Chuang, J.L, Gui, W.J, Wynn, R.M, Li, J, Chuang, D.T. | | Deposit date: | 2012-09-21 | | Release date: | 2013-06-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-based design and mechanisms of allosteric inhibitors for mitochondrial branched-chain alpha-ketoacid dehydrogenase kinase.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5T1P
 
 | | Crystal structure of the putative periplasmic solute-binding protein from Campylobacter jejuni | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ABC transporter, ... | | Authors: | Filippova, E.V, Wawrzsak, Z, Sandoval, J, Skarina, T, Grimshaw, S, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-08-19 | | Release date: | 2016-09-07 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the putative periplasmic solute-binding protein from Campylobacter jejuni
To Be Published
|
|
2HUW
 
 | |
3OTJ
 
 | | A Crystal Structure of Trypsin Complexed with BPTI (Bovine Pancreatic Trypsin Inhibitor) by X-ray/Neutron Joint Refinement | | Descriptor: | CALCIUM ION, Cationic trypsin, Pancreatic trypsin inhibitor, ... | | Authors: | Kawamura, K, Yamada, T, Kurihara, K, Tamada, T, Kuroki, R, Tanaka, I, Takahashi, H, Niimura, N. | | Deposit date: | 2010-09-12 | | Release date: | 2011-01-26 | | Last modified: | 2017-11-08 | | Method: | NEUTRON DIFFRACTION (2.15 Å), X-RAY DIFFRACTION | | Cite: | X-ray and neutron protein crystallographic analysis of the trypsin-BPTI complex.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
4I32
 
 | | Crystal structure of HCV NS3/4A D168V protease complexed with compound 4 | | Descriptor: | (2R,6S,7E,10E,13aR,14aR,16aS)-2-{[7-methoxy-8-methyl-2-(propan-2-yloxy)quinolin-4-yl]oxy}-N-[(1-methylcyclopropyl)sulfonyl]-6-{[(1-methyl-1H-pyrazol-3-yl)carbonyl]amino}-5,16-dioxo-1,2,3,6,9,12,13,13a,14,15,16,16a-dodecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecine-14a(5H)-carboxamide, Genome polyprotein, HCV non-structural protein 4A, ... | | Authors: | Lemke, C.T. | | Deposit date: | 2012-11-23 | | Release date: | 2013-01-02 | | Last modified: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (2.3001 Å) | | Cite: | Molecular Mechanism by Which a Potent Hepatitis C Virus NS3-NS4A Protease Inhibitor Overcomes Emergence of Resistance.
J.Biol.Chem., 288, 2013
|
|
4I33
 
 | | Crystal structure of HCV NS3/4A R155K protease complexed with compound 4 | | Descriptor: | (2R,6S,7E,10E,13aR,14aR,16aS)-2-{[7-methoxy-8-methyl-2-(propan-2-yloxy)quinolin-4-yl]oxy}-N-[(1-methylcyclopropyl)sulfonyl]-6-{[(1-methyl-1H-pyrazol-3-yl)carbonyl]amino}-5,16-dioxo-1,2,3,6,9,12,13,13a,14,15,16,16a-dodecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecine-14a(5H)-carboxamide, Genome polyprotein, HCV non-structural protein 4A, ... | | Authors: | Lemke, C.T. | | Deposit date: | 2012-11-23 | | Release date: | 2013-01-02 | | Last modified: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (1.9001 Å) | | Cite: | Molecular Mechanism by Which a Potent Hepatitis C Virus NS3-NS4A Protease Inhibitor Overcomes Emergence of Resistance.
J.Biol.Chem., 288, 2013
|
|
4KS5
 
 | | Influenza neuraminidase in complex with antiviral compound (3S,4R,5R)-4-(acetylamino)-3-[4-(2-hydroxypropan-2-yl)-1H-1,2,3-triazol-1-yl]-5-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid | | Descriptor: | (3S,4R,5R)-4-(acetylamino)-3-[4-(2-hydroxypropan-2-yl)-1H-1,2,3-triazol-1-yl]-5-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, CALCIUM ION, Neuraminidase | | Authors: | Kerry, P.S, Russell, R.J.M. | | Deposit date: | 2013-05-17 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.699 Å) | | Cite: | Structural basis for a class of nanomolar influenza A neuraminidase inhibitors.
Sci Rep, 3, 2013
|
|
4KZ6
 
 | | Crystal structure of AmpC beta-lactamase in complex with fragment 13 ((2R,6R)-6-methyl-1-(3-sulfanylpropanoyl)piperidine-2-carboxylic acid) | | Descriptor: | (2R,6R)-6-methyl-1-(3-sulfanylpropanoyl)piperidine-2-carboxylic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Eidam, O, Barelier, S, Fish, I, Shoichet, B.K. | | Deposit date: | 2013-05-29 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Increasing chemical space coverage by combining empirical and computational fragment screens.
Acs Chem.Biol., 9, 2014
|
|
2LFK
 
 | | NMR solution structure of native TdPI-short | | Descriptor: | Tryptase inhibitor | | Authors: | Bronsoms, S, Pantoja-Uceda, D, Gabrijelcic-Geiger, D, Sanglas, L, Aviles, F, Santoro, J, Sommerhoff, C, Arolas, J. | | Deposit date: | 2011-07-06 | | Release date: | 2011-11-09 | | Last modified: | 2023-12-06 | | Method: | SOLUTION NMR | | Cite: | Oxidative folding and structural analyses of a kunitz-related inhibitor and its disulfide intermediates: functional implications.
J.Mol.Biol., 414, 2011
|
|
2JBZ
 
 | | Crystal structure of the Streptomyces coelicolor holo-[Acyl-carrier-protein] Synthase (AcpS) in complex with coenzyme A at 1.6 A | | Descriptor: | COENZYME A, HOLO-[ACYL-CARRIER-PROTEIN] SYNTHASE, MAGNESIUM ION, ... | | Authors: | Dall'Aglio, P, Arthur, C, Law, C.K.E, Crump, M.P, Crosby, J, Hadfield, A.T. | | Deposit date: | 2006-12-15 | | Release date: | 2007-01-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Analysis of Streptomyces Coelicolor Phosphopantetheinyl Transferase, Acps, Reveals the Basis for Relaxed Substrate Specificity.
Biochemistry, 50, 2011
|
|
2L4O
 
 | | Solution structure of the Streptococcus pneumoniae RrgB pilus backbone D1 domain | | Descriptor: | Cell wall surface anchor family protein | | Authors: | Gentile, M.A, Melchiorre, S, Emolo, C, Moschioni, M, Gianfaldoni, C, Pancotto, L, Ferlenghi, I, Scarselli, M, Pansegrau, W, Veggi, D, Merola, M, Cantini, F, Ruggiero, P, Banci, L, Masignani, V. | | Deposit date: | 2010-10-11 | | Release date: | 2011-03-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Streptococcus pneumoniae RrgB pilus backbone D1 domain: in vivo protection and implications for the molecular mechanisms of pilus polymerization
To be Published
|
|
2LFL
 
 | | NMR solution structure of the intermediate IIIb of TdPI-short | | Descriptor: | Tryptase inhibitor | | Authors: | Bronsoms, S, Pantoja-Uceda, D, Gabrijelcic-Geiger, D, Sanglas, L, Aviles, F, Santoro, J, Sommerhoff, C, Arolas, J. | | Deposit date: | 2011-07-06 | | Release date: | 2011-11-09 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Oxidative folding and structural analyses of a kunitz-related inhibitor and its disulfide intermediates: functional implications.
J.Mol.Biol., 414, 2011
|
|
6AAZ
 
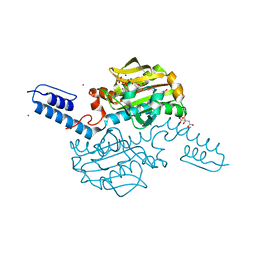 | | Crystal structure of Methanosarcina mazei PylRS(Y306A/Y384F) complexed with pNO2ZLys | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CITRIC ACID, MAGNESIUM ION, ... | | Authors: | Yanagisawa, T, Kuratani, M, Yokoyama, S. | | Deposit date: | 2018-07-19 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.842 Å) | | Cite: | Structural Basis for Genetic-Code Expansion with Bulky Lysine Derivatives by an Engineered Pyrrolysyl-tRNA Synthetase.
Cell Chem Biol, 26, 2019
|
|
6OOX
 
 | |
7ZYJ
 
 | |
6AB8
 
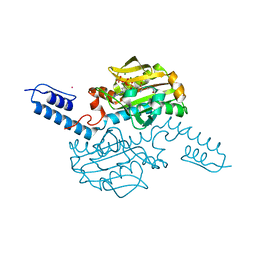 | | Crystal structure of Methanosarcina mazei PylRS(Y306A/Y384F) complexed with ZLys | | Descriptor: | (2S)-2-azanyl-6-(phenylmethoxycarbonylamino)hexanoic acid, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Yanagisawa, T, Kuratani, M, Yokoyama, S. | | Deposit date: | 2018-07-20 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.753 Å) | | Cite: | Structural Basis for Genetic-Code Expansion with Bulky Lysine Derivatives by an Engineered Pyrrolysyl-tRNA Synthetase.
Cell Chem Biol, 26, 2019
|
|
7ZX8
 
 | | Structure of SNAPc containing Pol II pre-initiation complex bound to U1 snRNA promoter (OC) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Rengachari, S, Schilbach, S, Kaliyappan, T, Gouge, J, Zumer, K, Schwarz, J, Urlaub, H, Dienemann, C, Vannini, A, Cramer, P. | | Deposit date: | 2022-05-20 | | Release date: | 2022-12-07 | | Last modified: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of SNAPc-dependent snRNA transcription initiation by RNA polymerase II.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6AB2
 
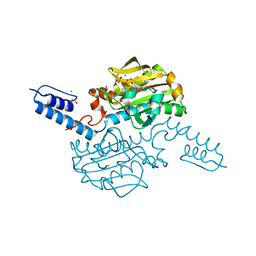 | | Crystal structure of Methanosarcina mazei PylRS(Y306A/Y384F) complexed with oClZLys | | Descriptor: | (2S)-2-azanyl-6-[(2-chlorophenyl)methoxycarbonylamino]hexanoic acid, 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Yanagisawa, T, Kuratani, M, Yokoyama, S. | | Deposit date: | 2018-07-19 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Basis for Genetic-Code Expansion with Bulky Lysine Derivatives by an Engineered Pyrrolysyl-tRNA Synthetase.
Cell Chem Biol, 26, 2019
|
|
2JWQ
 
 | | G-quadruplex recognition by quinacridines: a SAR, NMR and Biological study | | Descriptor: | DNA (5'-D(*DTP*DTP*DAP*DGP*DGP*DGP*DT)-3'), N,N'-(dibenzo[b,j][1,7]phenanthroline-2,10-diyldimethanediyl)dipropan-1-amine | | Authors: | Hounsou, C, Guittat, L, Monchaud, D, Jourdan, M, Saettel, N, Mergny, J.L, Teulade-Fichou, M. | | Deposit date: | 2007-10-23 | | Release date: | 2008-03-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | G-Quadruplex Recognition by Quinacridines: a SAR, NMR, and Biological Study
ChemMedChem, 2, 2007
|
|
