6CES
 
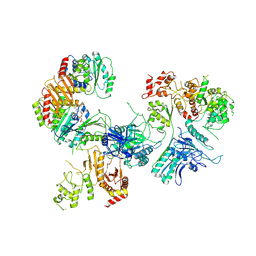 | | Cryo-EM structure of GATOR1-RAG | | Descriptor: | GATOR complex protein DEPDC5, GATOR complex protein NPRL2, GATOR complex protein NPRL3, ... | | Authors: | Shen, K, Huang, R.K, Brignole, E.J, Yu, Z, Sabatini, D.M. | | Deposit date: | 2018-02-12 | | Release date: | 2018-03-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Architecture of the human GATOR1 and GATOR1-Rag GTPases complexes.
Nature, 556, 2018
|
|
8T9D
 
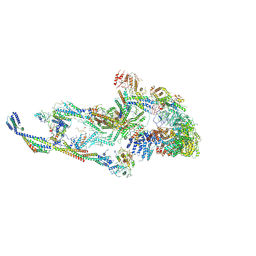 | | CryoEM structure of TR-TRAP | | Descriptor: | Mediator of RNA polymerase II transcription subunit 1, Mediator of RNA polymerase II transcription subunit 10, Mediator of RNA polymerase II transcription subunit 11, ... | | Authors: | Zhao, H, Asturias, F. | | Deposit date: | 2023-06-23 | | Release date: | 2024-07-03 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (4.66 Å) | | Cite: | An IDR-dependent mechanism for nuclear receptor control of Mediator interaction with RNA polymerase II.
Mol.Cell, 84, 2024
|
|
6D94
 
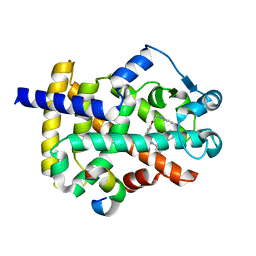 | | Crystal structure of PPAR gamma in complex with Mediator of RNA polymerase II transcription subunit 1 | | Descriptor: | (2~{S})-3-[4-[2-[methyl(pyridin-2-yl)amino]ethoxy]phenyl]-2-[[2-(phenylcarbonyl)phenyl]amino]propanoic acid, Mediator of RNA polymerase II transcription subunit 1, Peroxisome proliferator-activated receptor gamma | | Authors: | Mou, T.C, Chrisman, I.M, Hughes, T.S, Sprang, S.R. | | Deposit date: | 2018-04-27 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A structural mechanism of nuclear receptor biased agonism.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6DU3
 
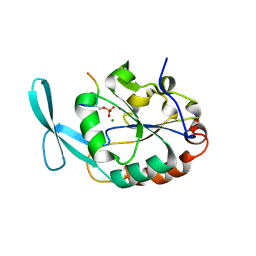 | | Structure of Scp1 D96N bound to REST-pS861/4 peptide | | Descriptor: | Carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1, MAGNESIUM ION, REST-pS861 | | Authors: | Burkholder, N.T, Mayfield, J.E, Yu, X, Irani, S, Arce, D.K, Jiang, F, Matthews, W, Xue, Y, Zhang, Y.J. | | Deposit date: | 2018-06-19 | | Release date: | 2018-09-26 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Phosphatase activity of small C-terminal domain phosphatase 1 (SCP1) controls the stability of the key neuronal regulator RE1-silencing transcription factor (REST).
J. Biol. Chem., 293, 2018
|
|
4RSX
 
 | |
4RSW
 
 | |
6ECN
 
 | | HIV-1 CA 1/2-hexamer-EE | | Descriptor: | HIV-1 CA | | Authors: | Summers, B.J, Xiong, Y. | | Deposit date: | 2018-08-08 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Modular HIV-1 Capsid Assemblies Reveal Diverse Host-Capsid Recognition Mechanisms.
Cell Host Microbe, 26, 2019
|
|
5JBE
 
 | | 4,6-alpha-glucanotransferase GTFB from Lactobacillus reuteri 121 complexed with an isomalto-maltopentasaccharide | | Descriptor: | ACETATE ION, CALCIUM ION, Inactive glucansucrase, ... | | Authors: | Pijning, T, Dijkstra, B.W, Bai, Y, Gangoiti-Munecas, J, Dijkhuizen, L. | | Deposit date: | 2016-04-13 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of 4,6-alpha-Glucanotransferase Supports Diet-Driven Evolution of GH70 Enzymes from alpha-Amylases in Oral Bacteria.
Structure, 25, 2017
|
|
5WIR
 
 | | Structure of the TRF1-TERB1 interface | | Descriptor: | TERB1-TBM, Telomeric repeat-binding factor 1 | | Authors: | Nandakumar, J, Pendlebury, D.F, Smith, E.M, Tesmer, V.M. | | Deposit date: | 2017-07-20 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dissecting the telomere-inner nuclear membrane interface formed in meiosis.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5JBF
 
 | | 4,6-alpha-glucanotransferase GTFB (D1015N mutant) from Lactobacillus reuteri 121 complexed with maltopentaose | | Descriptor: | CALCIUM ION, Inactive glucansucrase, SULFATE ION, ... | | Authors: | Pijning, T, Dijkstra, B.W, Bai, Y, Gangoiti-Munecas, J, Dijkhuizen, L. | | Deposit date: | 2016-04-13 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal Structure of 4,6-alpha-Glucanotransferase Supports Diet-Driven Evolution of GH70 Enzymes from alpha-Amylases in Oral Bacteria.
Structure, 25, 2017
|
|
6EC2
 
 | | Structure of HIV-1 CA 1/3-hexamer | | Descriptor: | ACETATE ION, Capsid protein p24 | | Authors: | Summers, B.J, Xiong, Y. | | Deposit date: | 2018-08-07 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Modular HIV-1 Capsid Assemblies Reveal Diverse Host-Capsid Recognition Mechanisms.
Cell Host Microbe, 26, 2019
|
|
6ECO
 
 | | Hexamer-2-Foldon HIV-1 capsid platform | | Descriptor: | HIV-1 capsid platform protein | | Authors: | Summers, B.J, Xiong, Y. | | Deposit date: | 2018-08-08 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Modular HIV-1 Capsid Assemblies Reveal Diverse Host-Capsid Recognition Mechanisms.
Cell Host Microbe, 26, 2019
|
|
6EFK
 
 | | Crystal structure of the human CHIP TPR domain in complex with a 5mer acetylated HSP70 peptide | | Descriptor: | ACE-ILE-GLU-GLU-VAL-ASP, E3 ubiquitin-protein ligase CHIP, SODIUM ION | | Authors: | Basu, K, Ravalin, M, Bohn, M.-F, Craik, C.S, Gestwicki, J.E. | | Deposit date: | 2018-08-16 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Specificity for latent C termini links the E3 ubiquitin ligase CHIP to caspases.
Nat.Chem.Biol., 15, 2019
|
|
4UI4
 
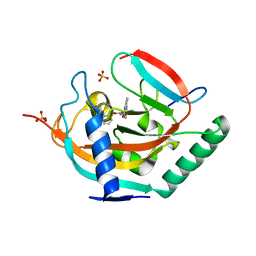 | | Crystal structure of human tankyrase 2 in complex with TA-29 | | Descriptor: | BENZYL N-{[4-(4-OXO-3,4-DIHYDROQUINAZOLIN-2-YL)PHENYL]METHYL}CARBAMATE, SULFATE ION, TANKYRASE-2, ... | | Authors: | Haikarainen, T, Lehtio, L. | | Deposit date: | 2015-03-27 | | Release date: | 2016-04-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Activity Relationships of 2-Arylquinazolin-4-Ones as Highly Selective and Potent Inhibitors of the Tankyrases.
Eur.J.Med.Chem., 118, 2016
|
|
4TXK
 
 | |
4UDP
 
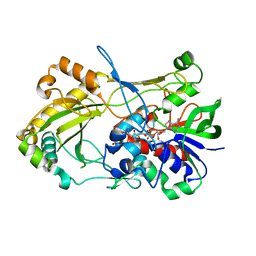 | | Crystal structure of 5-hydroxymethylfurfural oxidase (HMFO) in the oxidized state | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLUCOSE-METHANOL-CHOLINE OXIDOREDUCTASE | | Authors: | Dijkman, W, Binda, C, Fraaije, M, Mattevi, A. | | Deposit date: | 2014-12-11 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Based Enzyme Tailoring of 5-Hydroxymethylfurfural Oxidase
Acs Catalysis, 5, 2015
|
|
7KIF
 
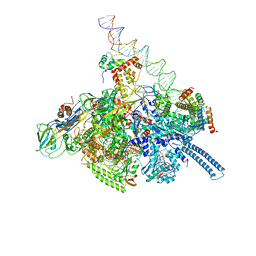 | | Mycobacterium tuberculosis WT RNAP transcription open promoter complex with WhiB7 transcription factor | | Descriptor: | DNA (55-MER), DNA (63-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Lilic, M, Darst, S.A, Campbell, E.A. | | Deposit date: | 2020-10-23 | | Release date: | 2021-04-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structural basis of transcriptional activation by the Mycobacterium tuberculosis intrinsic antibiotic-resistance transcription factor WhiB7.
Mol.Cell, 81, 2021
|
|
7KIM
 
 | | Mycobacterium tuberculosis WT RNAP transcription closed promoter complex with WhiB7 transcription factor | | Descriptor: | DNA (45-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Lilic, M, Darst, S.A, Campbell, E.A. | | Deposit date: | 2020-10-23 | | Release date: | 2021-04-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structural basis of transcriptional activation by the Mycobacterium tuberculosis intrinsic antibiotic-resistance transcription factor WhiB7.
Mol.Cell, 81, 2021
|
|
7KIN
 
 | | Mycobacterium tuberculosis WT RNAP transcription open promoter complex with WhiB7 promoter | | Descriptor: | DNA (49-MER), DNA (54-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Lilic, M, Darst, S.A, Campbell, E.A. | | Deposit date: | 2020-10-23 | | Release date: | 2021-04-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Structural basis of transcriptional activation by the Mycobacterium tuberculosis intrinsic antibiotic-resistance transcription factor WhiB7.
Mol.Cell, 81, 2021
|
|
4UDR
 
 | | Crystal structure of the H467A mutant of 5-hydroxymethylfurfural oxidase (HMFO) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FORMIC ACID, GLUCOSE-METHANOL-CHOLINE OXIDOREDUCTASE | | Authors: | Dijkman, W, Binda, C, Fraaije, M, Mattevi, A. | | Deposit date: | 2014-12-11 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Based Enzyme Tailoring of 5-Hydroxymethylfurfural Oxidase
Acs Catalysis, 5, 2015
|
|
4UDQ
 
 | | Crystal structure of 5-hydroxymethylfurfural oxidase (HMFO) in the reduced state | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FORMIC ACID, GLUCOSE-METHANOL-CHOLINE OXIDOREDUCTASE | | Authors: | Dijkman, W, Binda, C, Fraaije, M, Mattevi, A. | | Deposit date: | 2014-12-11 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Based Enzyme Tailoring of 5-Hydroxymethylfurfural Oxidase
Acs Catalysis, 5, 2015
|
|
5M0H
 
 | |
5M0N
 
 | | Crystal structure of cytochrome P450 OleT in complex with formate | | Descriptor: | FORMIC ACID, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ... | | Authors: | Tee, K.L, Munro, A, Matthews, S, Leys, D, Levy, C. | | Deposit date: | 2016-10-05 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Catalytic Determinants of Alkene Production by the Cytochrome P450 Peroxygenase OleTJE.
J. Biol. Chem., 292, 2017
|
|
5LJ5
 
 | | Overall structure of the yeast spliceosome immediately after branching. | | Descriptor: | CWC15, CWC22, Exon 1 (5' exon) of UBC4 pre-mRNA, ... | | Authors: | Galej, W.P, Wilkinson, M.F, Fica, S.M, Oubridge, C, Newman, A.J, Nagai, K. | | Deposit date: | 2016-07-17 | | Release date: | 2016-08-31 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of the spliceosome immediately after branching.
Nature, 537, 2016
|
|
5JBD
 
 | | 4,6-alpha-glucanotransferase GTFB from Lactobacillus reuteri 121 | | Descriptor: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Pijning, T, Dijkstra, B.W, Bai, Y, Gangoiti-Munecas, J, Dijkhuizen, L. | | Deposit date: | 2016-04-13 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of 4,6-alpha-Glucanotransferase Supports Diet-Driven Evolution of GH70 Enzymes from alpha-Amylases in Oral Bacteria.
Structure, 25, 2017
|
|
