4BRU
 
 | | Crystal structure of the yeast Dhh1-Edc3 complex | | Descriptor: | ATP-DEPENDENT RNA HELICASE DHH1, ENHANCER OF MRNA-DECAPPING PROTEIN 3 | | Authors: | Sharif, H, Ozgur, S, Sharma, K, Basquin, C, Urlaub, H, Conti, E. | | Deposit date: | 2013-06-05 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.245 Å) | | Cite: | Structural Analysis of the Yeast Dhh1-Pat1 Complex Reveals How Dhh1 Engages Pat1, Edc3 and RNA in Mutually Exclusive Interactions
Nucleic Acids Res., 41, 2013
|
|
6DJT
 
 | | Structure of TYW1 with a lysine-pyruvate adduct bound | | Descriptor: | DI(HYDROXYETHYL)ETHER, FE2/S3 CLUSTER, GLYCEROL, ... | | Authors: | Grell, T.A.J, Drennan, C.L. | | Deposit date: | 2018-05-26 | | Release date: | 2018-06-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Biochemical and Structural Characterization of a Schiff Base in the Radical-Mediated Biosynthesis of 4-Demethylwyosine by TYW1.
J. Am. Chem. Soc., 140, 2018
|
|
3FQK
 
 | | Hepatitis C virus polymerase NS5B (BK 1-570) with HCV-796 inhibitor | | Descriptor: | 5-cyclopropyl-2-(4-fluorophenyl)-6-[(2-hydroxyethyl)(methylsulfonyl)amino]-N-methyl-1-benzofuran-3-carboxamide, RNA-directed RNA polymerase | | Authors: | Harris, S.F, Wong, A. | | Deposit date: | 2009-01-07 | | Release date: | 2009-02-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Slow binding inhibition and mechanism of resistance of non-nucleoside polymerase inhibitors of hepatitis C virus.
J.Biol.Chem., 284, 2009
|
|
4MFF
 
 | | Structure of human DNA polymerase beta complexed with O6MG in the template base paired with incoming non-hydrolyzable TTP | | Descriptor: | 5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]thymidine, DNA polymerase beta, MAGNESIUM ION, ... | | Authors: | Koag, M.C, Min, K, Monzingo, A.F, Lee, S. | | Deposit date: | 2013-08-27 | | Release date: | 2014-04-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Metal-dependent conformational activation explains highly promutagenic replication across O6-methylguanine by human DNA polymerase beta.
J.Am.Chem.Soc., 136, 2014
|
|
5FQF
 
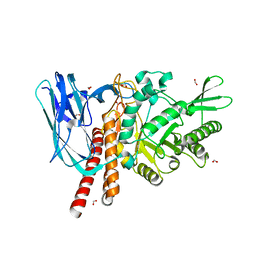 | | The details of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, BETA-N-ACETYLGALACTOSAMINIDASE, FORMIC ACID | | Authors: | Noach, I, Pluvinage, B, Laurie, C, Abe, K.T, Alteen, M, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2015-12-10 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Details of Glycolipid Glycan Hydrolysis by the Structural Analysis of a Family 123 Glycoside Hydrolase from Clostridium Perfringens
J.Mol.Biol., 428, 2016
|
|
5JCG
 
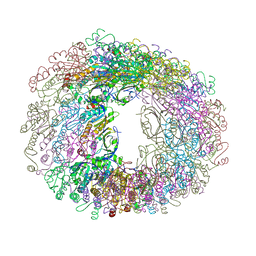 | |
1BUV
 
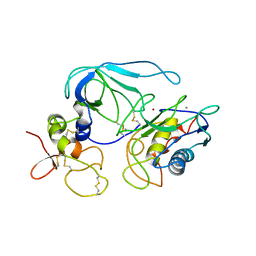 | | CRYSTAL STRUCTURE OF THE MT1-MMP-TIMP-2 COMPLEX | | Descriptor: | CALCIUM ION, PROTEIN (MEMBRANE-TYPE MATRIX METALLOPROTEINASE (CDMT1-MMP)), PROTEIN (METALLOPROTEINASE INHIBITOR (TIMP-2)), ... | | Authors: | Fernandez-Catalan, C, Bode, W, Huber, R, Turk, D, Calvete, J.J, Lichte, A, Tschesche, H, Maskos, K. | | Deposit date: | 1998-09-07 | | Release date: | 1999-09-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of the complex formed by the membrane type 1-matrix metalloproteinase with the tissue inhibitor of metalloproteinases-2, the soluble progelatinase A receptor.
EMBO J., 17, 1998
|
|
5FFR
 
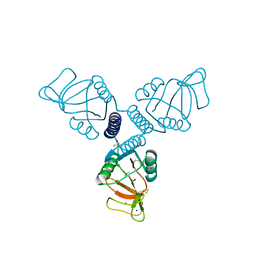 | | Crystal Structure of Surfactant Protein-A complexed with phosphocholine | | Descriptor: | CALCIUM ION, PHOSPHOCHOLINE, Pulmonary surfactant-associated protein A, ... | | Authors: | Goh, B.C, Wu, H, Rynkiewicz, M.J, Schulten, K, Seaton, B.A, McCormack, F.X. | | Deposit date: | 2015-12-18 | | Release date: | 2016-07-06 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Elucidation of Lipid Binding Sites on Lung Surfactant Protein A Using X-ray Crystallography, Mutagenesis, and Molecular Dynamics Simulations.
Biochemistry, 55, 2016
|
|
4MFC
 
 | | Structure of human DNA polymerase beta complexed with O6MG in the template base paired with incoming non-hydrolyzable CTP | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, DNA polymerase beta, MAGNESIUM ION, ... | | Authors: | Koag, M.C, Min, K, Monzingo, A.F, Lee, S. | | Deposit date: | 2013-08-27 | | Release date: | 2014-04-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Metal-dependent conformational activation explains highly promutagenic replication across O6-methylguanine by human DNA polymerase beta.
J.Am.Chem.Soc., 136, 2014
|
|
5J9H
 
 | |
1BXO
 
 | | ACID PROTEINASE (PENICILLOPEPSIN) (E.C.3.4.23.20) COMPLEX WITH PHOSPHONATE INHIBITOR: METHYL CYCLO[(2S)-2-[[(1R)-1-(N-(L-N-(3-METHYLBUTANOYL)VALYL-L-ASPARTYL)AMINO)-3-METHYLBUT YL] HYDROXYPHOSPHINYLOXY]-3-(3-AMINOMETHYL) PHENYLPROPANOATE | | Descriptor: | GLYCEROL, METHYL CYCLO[(2S)-2-[[(1R)-1-(N-(L-N-(3-METHYLBUTANOYL)VALYL-L-ASPARTYL)AMINO)-3-METHYLBUTYL]HYDROXYPHOSPHINYLOXY]-3-(3-AMINOMETHYL)PHENYLPROPANOATE, PROTEIN (PENICILLOPEPSIN), ... | | Authors: | Khan, A.R, Parrish, J.C, Fraser, M.E, Smith, W.W, Bartlett, P.A, James, M.N.G. | | Deposit date: | 1998-10-07 | | Release date: | 1998-10-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Lowering the entropic barrier for binding conformationally flexible inhibitors to enzymes.
Biochemistry, 37, 1998
|
|
5FFS
 
 | | Crystal Structure of Surfactant Protein-A Y164A Mutant | | Descriptor: | CALCIUM ION, Pulmonary surfactant-associated protein A | | Authors: | Goh, B.C, Wu, H, Rynkiewicz, M.J, Schulten, K, Seaton, B.A, McCormack, F.X. | | Deposit date: | 2015-12-18 | | Release date: | 2016-07-06 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Elucidation of Lipid Binding Sites on Lung Surfactant Protein A Using X-ray Crystallography, Mutagenesis, and Molecular Dynamics Simulations.
Biochemistry, 55, 2016
|
|
1BMD
 
 | |
4OOU
 
 | | Crystal structure of beta-1,4-D-mannanase from Cryptopygus antarcticus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-1,4-mannanase | | Authors: | Kim, M.-K, An, Y.J, Jeong, C.-S, Cha, S.-S. | | Deposit date: | 2014-02-04 | | Release date: | 2014-08-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structure-based investigation into the functional roles of the extended loop and substrate-recognition sites in an endo-beta-1,4-d-mannanase from the Antarctic springtail, Cryptopygus antarcticus.
Proteins, 82, 2014
|
|
3ZEU
 
 | | Structure of a Salmonella typhimurium YgjD-YeaZ heterodimer bound to ATPgammaS | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Nichols, C.E, Lamb, H.K, Thompson, P, El Omari, K, Lockyer, M, Charles, I, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2012-12-07 | | Release date: | 2013-03-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Crystal Structure of the Dimer of Two Essential Salmonella Typhimurium Proteins, Ygjd & Yeaz and Calorimetric Evidence for the Formation of a Ternary Ygjd-Yeaz-Yjee Complex.
Protein Sci., 22, 2013
|
|
6B2Y
 
 | | Apo YiuA Crystal Form 2 | | Descriptor: | SODIUM ION, Solute-binding periplasmic protein of iron/siderophore ABC transporter | | Authors: | Radka, C.D, DeLucas, L.J, Aller, S.G. | | Deposit date: | 2017-09-20 | | Release date: | 2017-11-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The crystal structure of the Yersinia pestis iron chaperone YiuA reveals a basic triad binding motif for the chelated metal.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
6CFW
 
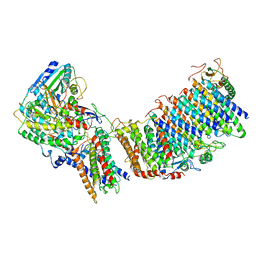 | |
1M0N
 
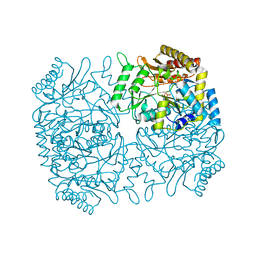 | | Structure of Dialkylglycine Decarboxylase Complexed with 1-Aminocyclopentanephosphonate | | Descriptor: | 1-[((1E)-{3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYLENE)AMINO]CYCLOPENTYLPHOSPHONIC ACID, 2,2-Dialkylglycine decarboxylase, POTASSIUM ION, ... | | Authors: | Liu, W, Rogers, C.J, Fisher, A.J, Toney, M.D. | | Deposit date: | 2002-06-13 | | Release date: | 2002-10-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Aminophosphonate Inhibitors of Dialkylglycine Decarboxylase: Structural Basis for Slow Binding Inhibition
Biochemistry, 41, 2002
|
|
1BLF
 
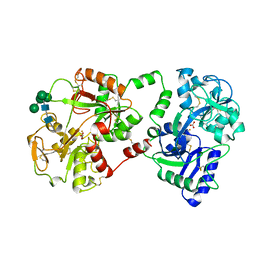 | | STRUCTURE OF DIFERRIC BOVINE LACTOFERRIN AT 2.8 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, FE (III) ION, ... | | Authors: | Moore, S.A, Anderson, B.F, Groom, C.R, Haridas, M, Baker, E.N. | | Deposit date: | 1997-08-20 | | Release date: | 1997-12-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Three-dimensional structure of diferric bovine lactoferrin at 2.8 A resolution.
J.Mol.Biol., 274, 1997
|
|
458D
 
 | |
6BFC
 
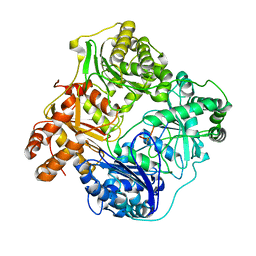 | | Cryo-EM structure of human insulin degrading enzyme in complex with insulin | | Descriptor: | Insulin, Insulin-degrading enzyme | | Authors: | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | Deposit date: | 2017-10-26 | | Release date: | 2017-12-27 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
6B3Q
 
 | | Cryo-EM structure of human insulin degrading enzyme in complex with insulin | | Descriptor: | Insulin, Insulin-degrading enzyme | | Authors: | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | Deposit date: | 2017-09-22 | | Release date: | 2017-11-22 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
4C5X
 
 | | Ultra High Resolution Dickerson-Drew dodecamer B-DNA with 5-Hydroxymethyl-cytosine Modification | | Descriptor: | 5'-D(CP*GP*CP*GP*AP*AP*TP*TP*5HCP*GP*CP*GP)-3', MAGNESIUM ION | | Authors: | McDonough, M, El-Sagheer, A.H, Brown, T, Schofield, C.J. | | Deposit date: | 2013-09-16 | | Release date: | 2013-10-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural insights into how 5-hydroxymethylation influences transcription factor binding.
Chem. Commun. (Camb.), 50, 2014
|
|
4GNF
 
 | | Crystal Structure of NSD3 tandem PHD5-C5HCH domains complexed with H3 peptide 1-15 | | Descriptor: | Histone H3.3, Histone-lysine N-methyltransferase NSD3, ZINC ION | | Authors: | Li, F, He, C, Wu, J, Shi, Y. | | Deposit date: | 2012-08-17 | | Release date: | 2013-01-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The methyltransferase NSD3 has chromatin-binding motifs, PHD5-C5HCH, that are distinct from other NSD (nuclear receptor SET domain) family members in their histone H3 recognition.
J.Biol.Chem., 288, 2013
|
|
4C63
 
 | | ULTRA HIGH RESOLUTION DICKERSON-DREW DODECAMER B-DNA WITH 5- METHYLCYSTOSINE MODIFICATION | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*5CMP*GP*CP*GP)-3', MAGNESIUM ION | | Authors: | Mcdonough, M, El-Sagheer, A.H, Brown, T, Schofield, C.J. | | Deposit date: | 2013-09-17 | | Release date: | 2013-10-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structural insights into how 5-hydroxymethylation influences transcription factor binding.
Chem. Commun. (Camb.), 50, 2014
|
|
