8E22
 
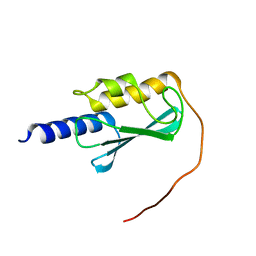 | | VPS37A_21-148 | | Descriptor: | Vacuolar protein sorting-associated protein 37A | | Authors: | Tian, F, Ye, Y.S. | | Deposit date: | 2022-08-12 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Identification of membrane curvature sensing motifs essential for VPS37A phagophore recruitment and autophagosome closure.
Commun Biol, 7, 2024
|
|
6O5Y
 
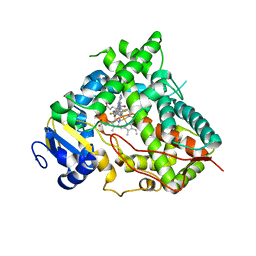 | | Structure of Human Cytochrome P450 1A1 with 5-amino-N-(5-((4R,5R)-4-amino-5-fluoroazepan-1-yl)-1-methyl-1H-pyrazol-4-yl)-2-(2,6-difluorophenyl)thiazole-4-carboxamide) | | Descriptor: | 5-amino-N-{5-[(4R,5R)-4-amino-5-fluoroazepan-1-yl]-1-methyl-1H-pyrazol-4-yl}-2-(2,6-difluorophenyl)-1,3-thiazole-4-carboxamide, Cytochrome P450 1A1, NITRATE ION, ... | | Authors: | Bart, A.G, Scott, E.E. | | Deposit date: | 2019-03-04 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Human Cytochrome P450 1A1 Adapts Active Site for Atypical Nonplanar Substrate.
Drug Metab.Dispos., 48, 2020
|
|
4B1M
 
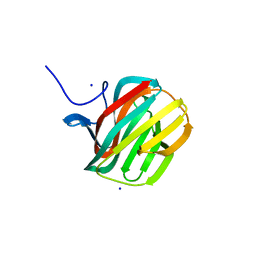 | | CARBOHYDRATE BINDING MODULE CBM66 FROM BACILLUS SUBTILIS | | Descriptor: | LEVANASE, SODIUM ION, SULFATE ION, ... | | Authors: | Cuskin, F, Flint, J.E, Morland, C, Basle, A, Henrissat, B, Countinho, P.M, Strazzulli, A, Solzehinkin, A, Davies, G.J, Gilbert, H.J, Gloster, T.M. | | Deposit date: | 2012-07-11 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | How Nature Can Exploit Nonspecific Catalytic and Carbohydrate Binding Modules to Create Enzymatic Specificity
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6KYY
 
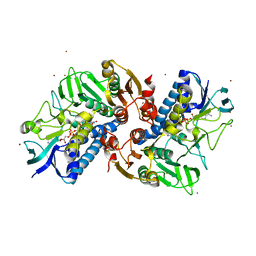 | |
4UFY
 
 | | Crystal structure of human tankyrase 2 in complex with TA-13 | | Descriptor: | 8-METHYL-2-(4-METHYLPHENYL)-3,4-DIHYDROQUINAZOLIN-4-ONE, GLYCEROL, SULFATE ION, ... | | Authors: | Haikarainen, T, Lehtio, L. | | Deposit date: | 2015-03-20 | | Release date: | 2016-04-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Activity Relationships of 2-Arylquinazolin-4-Ones as Highly Selective and Potent Inhibitors of the Tankyrases.
Eur.J.Med.Chem., 118, 2016
|
|
6L7O
 
 | | cryo-EM structure of cyanobacteria Fd-NDH-1L complex | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, BETA-CAROTENE, ... | | Authors: | Zhang, C, Shuai, J, Wu, J, Lei, M. | | Deposit date: | 2019-11-02 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into NDH-1 mediated cyclic electron transfer.
Nat Commun, 11, 2020
|
|
8EB3
 
 | |
4UIJ
 
 | | Crystal structure of the BTB domain of KCTD13 | | Descriptor: | BTB/POZ DOMAIN-CONTAINING ADAPTER FOR CUL3-MEDIATED RHOA DEGRADATION PROTEIN 1, CHLORIDE ION | | Authors: | Pinkas, D.M, Sanvitale, C.E, Sorell, F.J, Solcan, N, Goubin, S, Canning, P, Williams, E, Chaikuad, A, Dixon Clarke, S.E, Tallant, C, Fonseca, M, Chalk, R, Doutch, J, Krojer, T, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2015-03-30 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural complexity in the KCTD family of Cullin3-dependent E3 ubiquitin ligases.
Biochem. J., 474, 2017
|
|
4UI8
 
 | | Crystal structure of human tankyrase 2 in complex with TA-55 | | Descriptor: | 8-HYDROXY-2-[4-(TRIFLUOROMETHYL)PHENYL]-3,4-DIHYDROQUINAZOLIN-4-ONE, GLYCEROL, SULFATE ION, ... | | Authors: | Haikarainen, T, Lehtio, L. | | Deposit date: | 2015-03-27 | | Release date: | 2016-04-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-Activity Relationships of 2-Arylquinazolin-4-Ones as Highly Selective and Potent Inhibitors of the Tankyrases.
Eur.J.Med.Chem., 118, 2016
|
|
6LGD
 
 | | Bombyx mori GH13 sucrose hydrolase complexed with 1,4-dideoxy-1,4-imino-D-arabinitol | | Descriptor: | 1,4-DIDEOXY-1,4-IMINO-D-ARABINITOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Miyazaki, T. | | Deposit date: | 2019-12-05 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-function analysis of silkworm sucrose hydrolase uncovers the mechanism of substrate specificity in GH13 subfamily 17exo-alpha-glucosidases.
J.Biol.Chem., 295, 2020
|
|
4UI7
 
 | | Crystal structure of human tankyrase 2 in complex with TA-49 | | Descriptor: | 8-HYDROXY-2-(4-METHYLPHENYL)-3,4-DIHYDROQUINAZOLIN-4-ONE, GLYCEROL, SULFATE ION, ... | | Authors: | Haikarainen, T, Lehtio, L. | | Deposit date: | 2015-03-27 | | Release date: | 2016-04-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Activity Relationships of 2-Arylquinazolin-4-Ones as Highly Selective and Potent Inhibitors of the Tankyrases.
Eur.J.Med.Chem., 118, 2016
|
|
4WYK
 
 | |
6A60
 
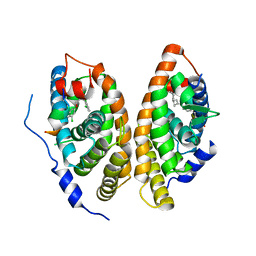 | | Crystal structure of human FXR/RXR-LBD heterodimer bound to GW4064 and 9cRA and SRC1 | | Descriptor: | (9cis)-retinoic acid, 3-[(E)-2-(2-chloro-4-{[3-(2,6-dichlorophenyl)-5-(1-methylethyl)isoxazol-4-yl]methoxy}phenyl)ethenyl]benzoic acid, Bile acid receptor, ... | | Authors: | Wang, N, Liu, J. | | Deposit date: | 2018-06-25 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Ligand binding and heterodimerization with retinoid X receptor alpha (RXR alpha ) induce farnesoid X receptor (FXR) conformational changes affecting coactivator binding
J. Biol. Chem., 293, 2018
|
|
6KYC
 
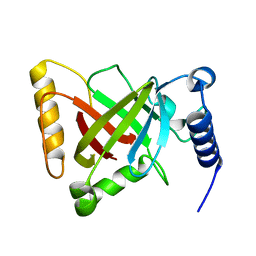 | | Structure of the S207A mutant of Clostridium difficile sortase B | | Descriptor: | Putative peptidase C60B, sortase B | | Authors: | Kang, C.Y, Huang, I.H, Wu, T.Y, Chang, J.C, Hsiao, Y.Y, Cheng, C.H, Tsai, W.J, Hsu, K.C, Wang, S.Y. | | Deposit date: | 2019-09-17 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Functional analysis ofClostridium difficilesortase B reveals key residues for catalytic activity and substrate specificity.
J.Biol.Chem., 295, 2020
|
|
8F0U
 
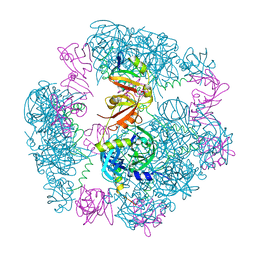 | | Structure of a 12mer DegP cage bound to the client protein hTRF1 | | Descriptor: | Periplasmic serine endoprotease DegP, Telomeric repeat-binding factor 1 | | Authors: | Harkness, R.W, Ripstein, Z.A, Di Trani, J.M, Kay, L.E. | | Deposit date: | 2022-11-04 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Flexible Client-Dependent Cages in the Assembly Landscape of the Periplasmic Protease-Chaperone DegP.
J.Am.Chem.Soc., 145, 2023
|
|
8F21
 
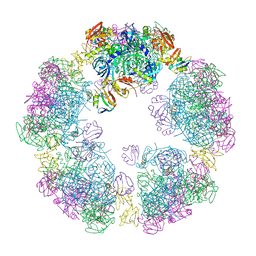 | | Structure of a 30mer DegP cage bound to the client protein hTRF1 | | Descriptor: | Periplasmic serine endoprotease DegP, Telomeric repeat-binding factor 1 | | Authors: | Harkness, R.W, Ripstein, Z.A, Di Trani, J.M, Kay, L.E. | | Deposit date: | 2022-11-06 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (14.1 Å) | | Cite: | Flexible Client-Dependent Cages in the Assembly Landscape of the Periplasmic Protease-Chaperone DegP.
J.Am.Chem.Soc., 145, 2023
|
|
8F0A
 
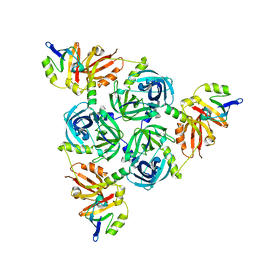 | | Client-bound structure of a DegP trimer within a 12mer cage | | Descriptor: | Periplasmic serine endoprotease DegP, Telomeric repeat-binding factor 1 | | Authors: | Harkness, R.W, Ripstein, Z.A, Di Trani, J.M, Kay, L.E. | | Deposit date: | 2022-11-02 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Flexible Client-Dependent Cages in the Assembly Landscape of the Periplasmic Protease-Chaperone DegP.
J.Am.Chem.Soc., 145, 2023
|
|
8F26
 
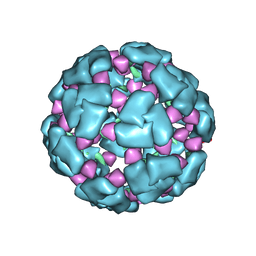 | | Structure of a 60mer DegP cage bound to the client protein hTRF1 | | Descriptor: | Periplasmic serine endoprotease DegP, Telomeric repeat-binding factor 1 | | Authors: | Harkness, R.W, Ripstein, Z.A, Di Trani, J.M, Kay, L.E. | | Deposit date: | 2022-11-07 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (9.7 Å) | | Cite: | Flexible Client-Dependent Cages in the Assembly Landscape of the Periplasmic Protease-Chaperone DegP.
J.Am.Chem.Soc., 145, 2023
|
|
4UF3
 
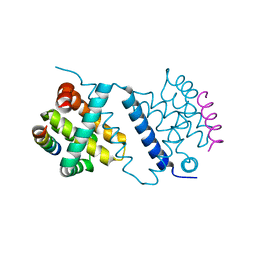 | | Deerpox virus DPV022 in complex with Bim BH3 | | Descriptor: | Antiapoptotic membrane protein, Bcl-2-like protein 11 | | Authors: | Burton, D.R, Kvansakul, M. | | Deposit date: | 2014-12-23 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis of Deerpox Virus-Mediated Inhibition of Apoptosis.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8F1T
 
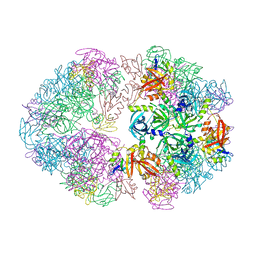 | | Structure of an 18mer DegP cage bound to the client protein hTRF1 | | Descriptor: | Periplasmic serine endoprotease DegP, Telomeric repeat-binding factor 1 | | Authors: | Harkness, R.W, Ripstein, Z.A, Di Trani, J.M, Kay, L.E. | | Deposit date: | 2022-11-06 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (12.1 Å) | | Cite: | Flexible Client-Dependent Cages in the Assembly Landscape of the Periplasmic Protease-Chaperone DegP.
J.Am.Chem.Soc., 145, 2023
|
|
8F1U
 
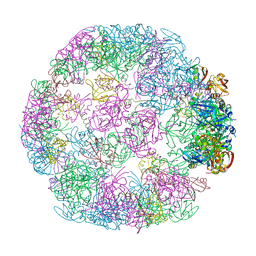 | | Structure of a 24mer DegP cage bound to the client protein hTRF1 | | Descriptor: | Periplasmic serine endoprotease DegP, Telomeric repeat-binding factor 1 | | Authors: | Harkness, R.W, Ripstein, Z.A, Di Trani, J.M, Kay, L.E. | | Deposit date: | 2022-11-06 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (13.8 Å) | | Cite: | Flexible Client-Dependent Cages in the Assembly Landscape of the Periplasmic Protease-Chaperone DegP.
J.Am.Chem.Soc., 145, 2023
|
|
6LE1
 
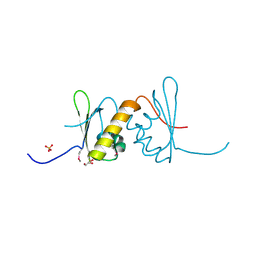 | | Structure of RRM2 domain of DND1 protein | | Descriptor: | Dead end protein homolog 1, GLYCEROL, SULFATE ION | | Authors: | Kumari, P, Bhavesh, N.S. | | Deposit date: | 2019-11-23 | | Release date: | 2020-12-02 | | Last modified: | 2021-06-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Human DND1-RRM2 forms a non-canonical domain swapped dimer.
Protein Sci., 30, 2021
|
|
8T9D
 
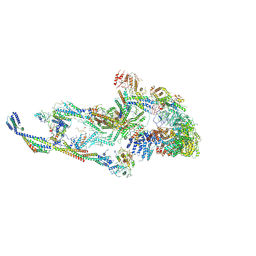 | | CryoEM structure of TR-TRAP | | Descriptor: | Mediator of RNA polymerase II transcription subunit 1, Mediator of RNA polymerase II transcription subunit 10, Mediator of RNA polymerase II transcription subunit 11, ... | | Authors: | Zhao, H, Asturias, F. | | Deposit date: | 2023-06-23 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.66 Å) | | Cite: | An IDR-dependent mechanism for nuclear receptor control of Mediator interaction with RNA polymerase II
To Be Published
|
|
4UI6
 
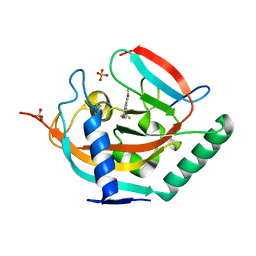 | | Crystal structure of human tankyrase 2 in complex with TA-47 | | Descriptor: | 8-METHOXY-2-[4-(TRIFLUOROMETHYL)PHENYL]-3,4-DIHYDROQUINAZOLIN-4-ONE, GLYCEROL, SULFATE ION, ... | | Authors: | Haikarainen, T, Lehtio, L. | | Deposit date: | 2015-03-27 | | Release date: | 2016-04-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Activity Relationships of 2-Arylquinazolin-4-Ones as Highly Selective and Potent Inhibitors of the Tankyrases.
Eur.J.Med.Chem., 118, 2016
|
|
4UI3
 
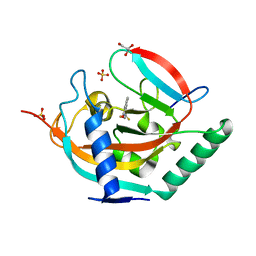 | | Crystal structure of human tankyrase 2 in complex with TA-26 | | Descriptor: | 2-[4-(trifluoromethyl)phenyl]-3H-quinazolin-4-one, GLYCEROL, SULFATE ION, ... | | Authors: | Haikarainen, T, Lehtio, L. | | Deposit date: | 2015-03-27 | | Release date: | 2016-04-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Activity Relationships of 2-Arylquinazolin-4-Ones as Highly Selective and Potent Inhibitors of the Tankyrases.
Eur.J.Med.Chem., 118, 2016
|
|
