7KDU
 
 | | Ricin bound to VHH antibody V5E4 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ricin chain A, ... | | Authors: | Rudolph, M.J. | | Deposit date: | 2020-10-09 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.809 Å) | | Cite: | Structural Analysis of Toxin-Neutralizing, Single-Domain Antibodies that Bridge Ricin's A-B Subunit Interface.
J.Mol.Biol., 433, 2021
|
|
7KBK
 
 | | Ricin bound to VHH antibody V6E11 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Ricin, ... | | Authors: | Rudolph, M.J. | | Deposit date: | 2020-10-02 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.091 Å) | | Cite: | Structural Analysis of Toxin-Neutralizing, Single-Domain Antibodies that Bridge Ricin's A-B Subunit Interface.
J.Mol.Biol., 433, 2021
|
|
7KDM
 
 | | Ricin bound to VHH antibody V5G6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Anti-RON nanobody, CHLORIDE ION, ... | | Authors: | Rudolph, M.J. | | Deposit date: | 2020-10-08 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structural Analysis of Toxin-Neutralizing, Single-Domain Antibodies that Bridge Ricin's A-B Subunit Interface.
J.Mol.Biol., 433, 2021
|
|
7KC9
 
 | | Ricin bound to VHH antibody V5G1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Ricin chain A, ... | | Authors: | Rudolph, M.J. | | Deposit date: | 2020-10-05 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structural Analysis of Toxin-Neutralizing, Single-Domain Antibodies that Bridge Ricin's A-B Subunit Interface.
J.Mol.Biol., 433, 2021
|
|
7KBI
 
 | | Ricin bound to VHH antibody V5E1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Ricin chain A, ... | | Authors: | Rudolph, M.J. | | Deposit date: | 2020-10-02 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.049 Å) | | Cite: | Structural Analysis of Toxin-Neutralizing, Single-Domain Antibodies that Bridge Ricin's A-B Subunit Interface.
J.Mol.Biol., 433, 2021
|
|
5K3Q
 
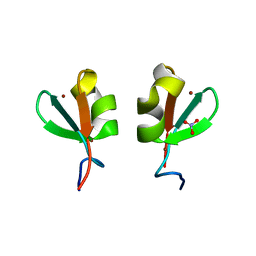 | | Rhesus macaques Trim5alpha Bbox2 domain | | Descriptor: | NITRATE ION, Tripartite motif-containing protein 5,Tripartite motif-containing protein 5, ZINC ION | | Authors: | Keown, J.K, Goldstone, D.C. | | Deposit date: | 2016-05-19 | | Release date: | 2016-07-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the Trim5 alpha Bbox2 domain from rhesus macaques describes a plastic oligomerisation interface.
J.Struct.Biol., 195, 2016
|
|
1X3M
 
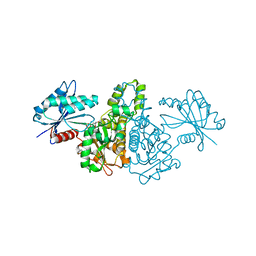 | |
1X3N
 
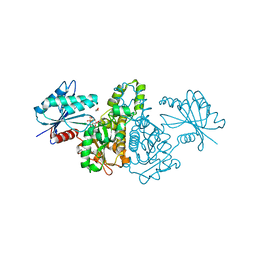 | | Crystal structure of AMPPNP bound Propionate kinase (TdcD) from Salmonella typhimurium | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Propionate kinase | | Authors: | Simanshu, D.K, Savithri, H.S, Murthy, M.R. | | Deposit date: | 2005-05-10 | | Release date: | 2005-09-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of ADP and AMPPNP-bound Propionate Kinase (TdcD) from Salmonella typhimurium: Comparison with Members of Acetate and Sugar Kinase/Heat Shock Cognate 70/Actin Superfamily.
J.Mol.Biol., 352, 2005
|
|
6GE4
 
 | |
6GE5
 
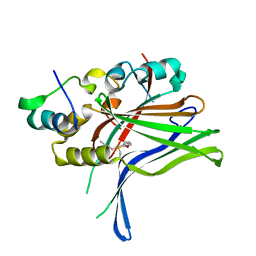 | |
6GEC
 
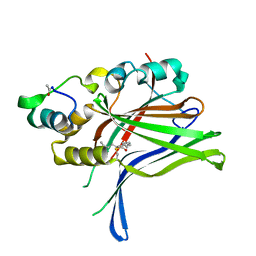 | |
6GEI
 
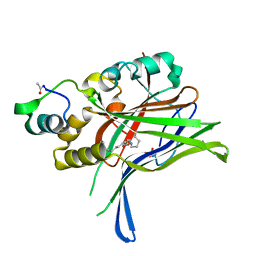 | |
6GEG
 
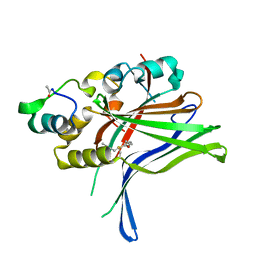 | |
6GEK
 
 | |
6GEE
 
 | |
6OIZ
 
 | | Amyloid-Beta (20-34) wild type | | Descriptor: | Amyloid beta A4 protein | | Authors: | Sawaya, M.R, Warmack, R.A, Zee, C.T, Gonen, T, Clarke, S.G, Eisenberg, D.S. | | Deposit date: | 2019-04-10 | | Release date: | 2019-08-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.1 Å) | | Cite: | Structure of amyloid-beta (20-34) with Alzheimer's-associated isomerization at Asp23 reveals a distinct protofilament interface.
Nat Commun, 10, 2019
|
|
3I6B
 
 | |
3HYC
 
 | |
2BGH
 
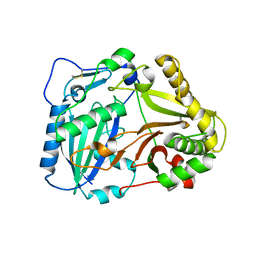 | | Crystal structure of Vinorine Synthase | | Descriptor: | VINORINE SYNTHASE | | Authors: | Ma, X, Koepke, J, Panjikar, S, Fritzsch, G, Stoeckigt, J. | | Deposit date: | 2004-12-22 | | Release date: | 2005-01-24 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Vinorine Synthase, the First Representative of the Bahd Superfamily.
J.Biol.Chem., 280, 2005
|
|
5IZ2
 
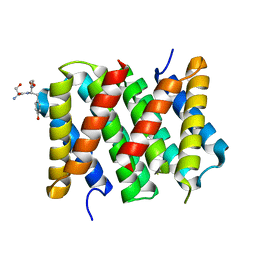 | | Crystal structure of the N. clavipes spidroin NTD at pH 6.5 | | Descriptor: | Major ampullate spidroin 1A, Major ampullate spidroin 1A (Partial C-terminus) | | Authors: | Atkison, J.H, Olsen, S.K. | | Deposit date: | 2016-03-24 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal Structure of the Nephila clavipes Major Ampullate Spidroin 1A N-terminal Domain Reveals Plasticity at the Dimer Interface.
J.Biol.Chem., 291, 2016
|
|
7A8X
 
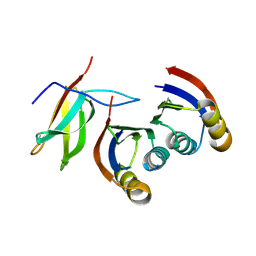 | | Complex of rice blast (Magnaporthe oryzae) effector protein AVR-PikC with the HMA domain of Pikh-1 from rice (Oryza sativa) | | Descriptor: | AVR-Pik protein, NBS-LRR class disease resistance protein | | Authors: | Maidment, J.H.R, Xiao, G, Franceschetti, M, Banfield, M.J. | | Deposit date: | 2020-08-31 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The allelic rice immune receptor Pikh confers extended resistance to strains of the blast fungus through a single polymorphism in the effector binding interface.
Plos Pathog., 17, 2021
|
|
5BXX
 
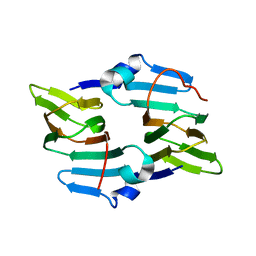 | | Crystal structure of the ectoine synthase from the cold-adapted marine bacterium Sphingopyxis alaskensis | | Descriptor: | L-ectoine synthase | | Authors: | Widderich, N, Kobus, S, Hoeppner, A, Bremer, E, Smits, S.H.J. | | Deposit date: | 2015-06-09 | | Release date: | 2016-04-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemistry and Crystal Structure of Ectoine Synthase: A Metal-Containing Member of the Cupin Superfamily.
Plos One, 11, 2016
|
|
4PU0
 
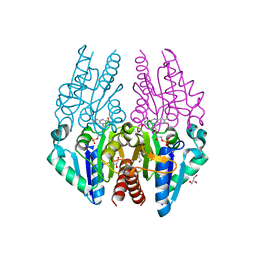 | | Crystal structure of the Escherichia coli alkanesulfonate FMN reductase SsuE in FMNH2-bound form | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN reductase SsuE, GLYCEROL, ... | | Authors: | Driggers, C.M, Ellis, H.R, Karplus, P.A. | | Deposit date: | 2014-03-11 | | Release date: | 2014-06-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3018 Å) | | Cite: | Crystal Structure of Escherichia coli SsuE: Defining a General Catalytic Cycle for FMN Reductases of the Flavodoxin-like Superfamily.
Biochemistry, 53, 2014
|
|
5BY5
 
 | | High resolution structure of the ectoine synthase from the cold-adapted marine bacterium Sphingopyxis alaskensis | | Descriptor: | L-ectoine synthase, S-1,2-PROPANEDIOL | | Authors: | Widderich, N, Kobus, S, Hoeppner, A, Bremer, E, Smits, S.H.J. | | Deposit date: | 2015-06-10 | | Release date: | 2016-04-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Biochemistry and Crystal Structure of Ectoine Synthase: A Metal-Containing Member of the Cupin Superfamily.
Plos One, 11, 2016
|
|
4PTZ
 
 | | Crystal structure of the Escherichia coli alkanesulfonate FMN reductase SsuE in FMN-bound form | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN reductase SsuE, GLYCEROL, ... | | Authors: | Driggers, C.M, Ellis, H.R, Karplus, P.A. | | Deposit date: | 2014-03-11 | | Release date: | 2014-06-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9007 Å) | | Cite: | Crystal Structure of Escherichia coli SsuE: Defining a General Catalytic Cycle for FMN Reductases of the Flavodoxin-like Superfamily.
Biochemistry, 53, 2014
|
|
