1AKT
 
 | | G61N OXIDIZED FLAVODOXIN MUTANT | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Mccarthy, A, Walsh, M, Higgins, T. | | Deposit date: | 1997-05-27 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modulation of the redox potentials of FMN in Desulfovibrio vulgaris flavodoxin: thermodynamic properties and crystal structures of glycine-61 mutants.
Biochemistry, 37, 1998
|
|
1AKU
 
 | | D95A HYDROQUINONE FLAVODOXIN MUTANT FROM D. VULGARIS | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN, SULFATE ION | | Authors: | Mccarthy, A, Walsh, M, Higgins, T. | | Deposit date: | 1997-05-27 | | Release date: | 1998-12-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic investigation of the role of aspartate 95 in the modulation of the redox potentials of Desulfovibrio vulgaris flavodoxin.
Biochemistry, 41, 2002
|
|
1AKV
 
 | | D95A SEMIQUINONE FLAVODOXIN MUTANT FROM D. VULGARIS | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN, SULFATE ION | | Authors: | Mccarthy, A, Walsh, M, Higgins, T. | | Deposit date: | 1997-05-27 | | Release date: | 1998-12-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic investigation of the role of aspartate 95 in the modulation of the redox potentials of Desulfovibrio vulgaris flavodoxin.
Biochemistry, 41, 2002
|
|
1AKW
 
 | | G61L OXIDIZED FLAVODOXIN MUTANT | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Mccarthy, A, Walsh, M, Higgins, T. | | Deposit date: | 1997-05-27 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Modulation of the redox potentials of FMN in Desulfovibrio vulgaris flavodoxin: thermodynamic properties and crystal structures of glycine-61 mutants.
Biochemistry, 37, 1998
|
|
1AKX
 
 | |
1AKY
 
 | |
1AKZ
 
 | | HUMAN URACIL-DNA GLYCOSYLASE | | Descriptor: | URACIL-DNA GLYCOSYLASE | | Authors: | Tainer, J.A, Mol, C.D. | | Deposit date: | 1997-05-27 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA.
Embo J., 17, 1998
|
|
1AL0
 
 | | PROCAPSID OF BACTERIOPHAGE PHIX174 | | Descriptor: | CAPSID PROTEIN GPF, SCAFFOLDING PROTEIN GPB, SCAFFOLDING PROTEIN GPD, ... | | Authors: | Rossmann, M.G, Dokland, T. | | Deposit date: | 1997-06-06 | | Release date: | 1998-01-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of a viral procapsid with molecular scaffolding.
Nature, 389, 1997
|
|
1AL1
 
 | | CRYSTAL STRUCTURE OF ALPHA1: IMPLICATIONS FOR PROTEIN DESIGN | | Descriptor: | ALPHA HELIX PEPTIDE: ELLKKLLEELKG, SULFATE ION | | Authors: | Hill, C.P, Anderson, D.H, Wesson, L, Degrado, W.F, Eisenberg, D. | | Deposit date: | 1990-07-02 | | Release date: | 1991-10-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of alpha 1: implications for protein design.
Science, 249, 1990
|
|
1AL2
 
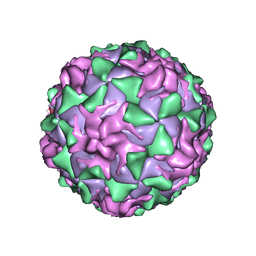 | | P1/MAHONEY POLIOVIRUS, SINGLE SITE MUTANT V1160I | | Descriptor: | MYRISTIC ACID, P1/MAHONEY POLIOVIRUS, SPHINGOSINE | | Authors: | Wien, M.W, Curry, S, Filman, D.J, Hogle, J.M. | | Deposit date: | 1997-06-09 | | Release date: | 1997-11-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural studies of poliovirus mutants that overcome receptor defects.
Nat.Struct.Biol., 4, 1997
|
|
1AL3
 
 | |
1AL4
 
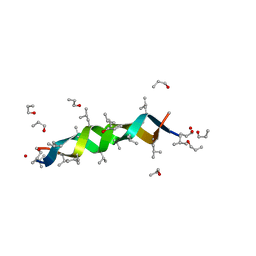 | | GRAMICIDIN D FROM BACILLUS BREVIS (N-PROPANOL SOLVATE) | | Descriptor: | GRAMICIDIN D, N-PROPANOL | | Authors: | Burkhart, B.M, Gassman, R.M, Pangborn, W.A, Duax, W.L. | | Deposit date: | 1997-06-11 | | Release date: | 1998-03-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Heterodimer Formation and Crystal Nucleation of Gramicidin D
Biophys.J., 75, 1998
|
|
1AL5
 
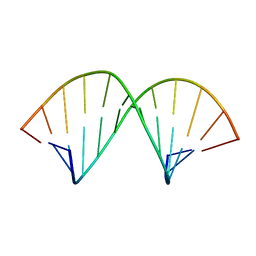 | | A-TRACT RNA DODECAMER, NMR, 12 STRUCTURES | | Descriptor: | RNA (5'-R(*CP*GP*CP*AP*AP*AP*UP*UP*UP*GP*CP*G)-3') | | Authors: | Conte, M.R, Conn, G.L, Brown, T, Lane, A.N. | | Deposit date: | 1997-06-11 | | Release date: | 1997-12-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Conformational properties and thermodynamics of the RNA duplex r(CGCAAAUUUGCG)2: comparison with the DNA analogue d(CGCAAATTTGCG)2.
Nucleic Acids Res., 25, 1997
|
|
1AL6
 
 | |
1AL7
 
 | |
1AL8
 
 | |
1AL9
 
 | |
1ALA
 
 | |
1ALB
 
 | |
1ALC
 
 | |
1ALD
 
 | |
1ALE
 
 | |
1ALF
 
 | |
1ALG
 
 | | SOLUTION STRUCTURE OF AN HGR INHIBITOR, NMR, 10 STRUCTURES | | Descriptor: | P11 | | Authors: | Schott, M.K, Nordhoff, A, Becker, K, Kalbitzer, H.R, Schirmer, R.H. | | Deposit date: | 1997-06-03 | | Release date: | 1997-10-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Denaturation and reactivation of dimeric human glutathione reductase--an assay for folding inhibitors.
Eur.J.Biochem., 245, 1997
|
|
1ALH
 
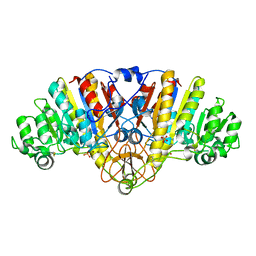 | | KINETICS AND CRYSTAL STRUCTURE OF A MUTANT E. COLI ALKALINE PHOSPHATASE (ASP-369-->ASN): A MECHANISM INVOLVING ONE ZINC PER ACTIVE SITE | | Descriptor: | ALKALINE PHOSPHATASE, PHOSPHATE ION, SULFATE ION, ... | | Authors: | Tibbitts, T.T, Xu, X, Kantrowitz, E.R. | | Deposit date: | 1994-08-23 | | Release date: | 1995-02-27 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Kinetics and crystal structure of a mutant Escherichia coli alkaline phosphatase (Asp-369-->Asn): a mechanism involving one zinc per active site.
Protein Sci., 3, 1994
|
|
