3FFG
 
 | | Factor XA in complex with the inhibitor (R)-6-(2'-((3- HYDROXYPYRROLIDIN-1-YL)METHYL)BIPHENYL-4-YL)-1-(3-(5-OXO-4,5-DIHYDRO-1H-1,2,4-TRIAZOL-3-YL)PHENYL)-3-(TRIFLUOROMETHYL)-5,6-DIHYDRO-1H-PYRAZOLO[3,4-C]PYRIDIN- 7(4H)-ONE | | Descriptor: | (R)-6-(2'-((3-hydroxypyrrolidin-1-yl)methyl)biphenyl-4-yl)-1-(3-(5-oxo-4,5-dihydro-1h-1,2,4-triazol-3-yl)phenyl)-3-(trifluoromethyl)-5,6-dihydro-1h-pyrazolo[3,4-c]pyridin-7(4h)-one, Coagulation factor X, heavy chain, ... | | Authors: | Alexander, R.A. | | Deposit date: | 2008-12-03 | | Release date: | 2009-12-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Phenyltriazolinones as potent factor Xa inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3F64
 
 | | F17a-G lectin domain with bound GlcNAc(beta1-O)paranitrophenyl ligand | | Descriptor: | 4-nitrophenyl 2-acetamido-2-deoxy-beta-D-glucopyranoside, F17a-G | | Authors: | Buts, L, De Boer, A, Olsson, J.D.M, Jonckheere, W, De Kerpel, M, De Genst, E, Guerardel, Y, Willaert, R, Wyns, L, Wuhrer, M, Oscarson, S, De Greve, H, Bouckaert, J. | | Deposit date: | 2008-11-05 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Sampling of Glycan Interaction Profiles Reveals Mucosal Receptors for Fimbrial Adhesins of Enterotoxigenic Escherichia coli.
Biology (Basel), 2, 2013
|
|
6IUL
 
 | |
3F9P
 
 | | Crystal structure of myeloperoxidase from human leukocytes | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CALCIUM ION, ... | | Authors: | Carpena, X, Fita, I, Obinger, C. | | Deposit date: | 2008-11-14 | | Release date: | 2009-07-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Essential role of proximal histidine-asparagine interaction in Mammalian peroxidases.
J.Biol.Chem., 284, 2009
|
|
6IMB
 
 | | Crystal structure of PDE4D complexed with a novel inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 6,7-dimethoxy-3,4-dihydroisoquinoline-2(1H)-carbaldehyde, MAGNESIUM ION, ... | | Authors: | Zhang, X, Su, H, Xu, Y. | | Deposit date: | 2018-10-22 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | Structure-Aided Identification and Optimization of Tetrahydro-isoquinolines as Novel PDE4 Inhibitors Leading to Discovery of an Effective Antipsoriasis Agent.
J.Med.Chem., 62, 2019
|
|
6IMI
 
 | | Crystal structure of PDE4D complexed with a novel inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 6-ethoxy-7-methoxy-3,4-dihydroisoquinoline-2(1H)-carbaldehyde, MAGNESIUM ION, ... | | Authors: | Zhang, X, Su, H, Xu, Y. | | Deposit date: | 2018-10-23 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structure-Aided Identification and Optimization of Tetrahydro-isoquinolines as Novel PDE4 Inhibitors Leading to Discovery of an Effective Antipsoriasis Agent.
J.Med.Chem., 62, 2019
|
|
1MAJ
 
 | | SOLUTION STRUCTURE OF AN ISOLATED ANTIBODY VL DOMAIN | | Descriptor: | IGG2A-KAPPA 26-10 FV (LIGHT CHAIN) | | Authors: | Constantine, K.L, Friedrichs, M.S, Metzler, W.J, Wittekind, M, Hensley, P, Mueller, L. | | Deposit date: | 1993-09-16 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an isolated antibody VL domain.
J.Mol.Biol., 236, 1994
|
|
3H5S
 
 | | Hepatitis C virus polymerase NS5B with saccharin inhibitor | | Descriptor: | (5S)-5-tert-butyl-1-(4-fluoro-3-methylbenzyl)-4-hydroxy-3-[8-(methylsulfonyl)-1,1-dioxido-6,7,8,9-tetrahydroisothiazolo[4,5-h]isoquinolin-3-yl]-1,5-dihydro-2H-pyrrol-2-one, RNA-directed RNA polymerase | | Authors: | Harris, S.F, Wong, A. | | Deposit date: | 2009-04-22 | | Release date: | 2009-09-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Non-nucleoside inhibitors of HCV polymerase NS5B. Part 4: structure-based design, synthesis, and biological evaluation of benzo[d]isothiazole-1,1-dioxides
Bioorg.Med.Chem.Lett., 19, 2009
|
|
6LLE
 
 | | CryoEM structure of SERCA2b WT in E1-2Ca2+-AMPPCP state. | | Descriptor: | CALCIUM ION, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Zhang, Y, Tsutsumi, A, Watanabe, S, Inaba, K. | | Deposit date: | 2019-12-23 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of SERCA2b reveal the mechanism of regulation by the luminal extension tail.
Sci Adv, 6, 2020
|
|
6IMD
 
 | | Crystal structure of PDE4D complexed with a novel inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 6,7-dimethoxy-3,4-dihydroisoquinoline-2(1H)-carbaldehyde, MAGNESIUM ION, ... | | Authors: | Zhang, X, Su, H, Xu, Y. | | Deposit date: | 2018-10-22 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Structure-Aided Identification and Optimization of Tetrahydro-isoquinolines as Novel PDE4 Inhibitors Leading to Discovery of an Effective Antipsoriasis Agent.
J.Med.Chem., 62, 2019
|
|
3GYJ
 
 | |
3GOK
 
 | | Binding site mapping of protein ligands | | Descriptor: | 2-(2-QUINOLIN-3-YLPYRIDIN-4-YL)-1,5,6,7-TETRAHYDRO-4H-PYRROLO[3,2-C]PYRIDIN-4-ONE, MAP kinase-activated protein kinase 2 | | Authors: | Scheich, C. | | Deposit date: | 2009-03-19 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Binding site mapping of protein ligands
To be published
|
|
3H96
 
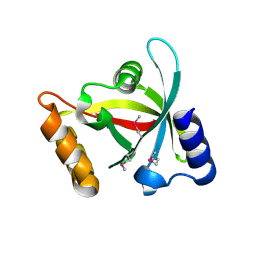 | | Msmeg_3358 F420 Reductase | | Descriptor: | 1,2-ETHANEDIOL, F420-H2 Dependent Reductase A | | Authors: | Jackson, C.J, French, N, Newman, J, Taylor, M.C, Russell, R.J, Oakeshott, J.G. | | Deposit date: | 2009-04-30 | | Release date: | 2010-04-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification and characterization of two families of F420 H2-dependent reductases from Mycobacteria that catalyse aflatoxin degradation.
Mol.Microbiol., 78, 2010
|
|
1QHF
 
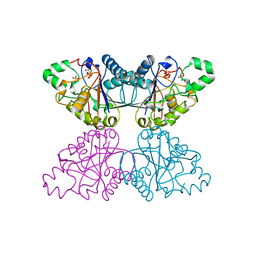 | | YEAST PHOSPHOGLYCERATE MUTASE-3PG COMPLEX STRUCTURE TO 1.7 A | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, PROTEIN (PHOSPHOGLYCERATE MUTASE), SULFATE ION | | Authors: | Crowhurst, G, Littlechild, J, Watson, H.C. | | Deposit date: | 1999-05-13 | | Release date: | 1999-06-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a phosphoglycerate mutase:3-phosphoglyceric acid complex at 1.7 A.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1MAK
 
 | | SOLUTION STRUCTURE OF AN ISOLATED ANTIBODY VL DOMAIN | | Descriptor: | IGG2A-KAPPA 26-10 FV (LIGHT CHAIN) | | Authors: | Constantine, K.L, Friedrichs, M.S, Metzler, W.J, Wittekind, M, Hensley, P, Mueller, L. | | Deposit date: | 1993-09-16 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an isolated antibody VL domain.
J.Mol.Biol., 236, 1994
|
|
1QMY
 
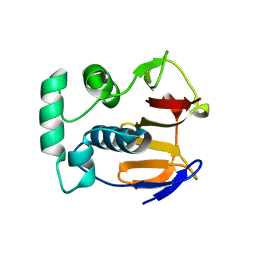 | | FMDV LEADER PROTEASE (LBSHORT-C51A-C133S) | | Descriptor: | 1,2-ETHANEDIOL, PROTEASE | | Authors: | Guarne, A, Tormo, J, Glaser, W, Skern, T, Fita, I. | | Deposit date: | 1999-10-08 | | Release date: | 2000-10-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Biochemical Features Distinguish the Foot-and-Mouth Disease Virus Leader Proteinase from Other Papain-Like Enzymes
J.Mol.Biol., 302, 2000
|
|
6IM6
 
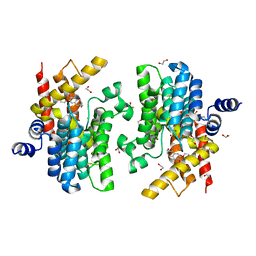 | | Crystal structure of PDE4D complexed with a novel inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 7-ethoxy-6-methoxy-3,4-dihydroisoquinoline-2(1H)-carbaldehyde, MAGNESIUM ION, ... | | Authors: | Zhang, X.L, Su, H.X, Xu, Y.C. | | Deposit date: | 2018-10-22 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Structure-Aided Identification and Optimization of Tetrahydro-isoquinolines as Novel PDE4 Inhibitors Leading to Discovery of an Effective Antipsoriasis Agent.
J.Med.Chem., 62, 2019
|
|
6IMT
 
 | | Crystal structure of PDE4D complexed with a novel inhibitor | | Descriptor: | (1S)-1-[2-(6-fluoro-1H-indol-3-yl)ethyl]-6,7-dimethoxy-3,4-dihydroisoquinoline-2(1H)-carbaldehyde, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Zhang, X.L, Su, H.X, Xu, Y.C. | | Deposit date: | 2018-10-23 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.483 Å) | | Cite: | Structure-Aided Identification and Optimization of Tetrahydro-isoquinolines as Novel PDE4 Inhibitors Leading to Discovery of an Effective Antipsoriasis Agent.
J.Med.Chem., 62, 2019
|
|
3FZC
 
 | | OXA-24 beta-lactamase complex with SA3-53 inhibitor | | Descriptor: | (2S,3R)-4-(2-aminoethylcarbamoyloxy)-2-[(2-methanoylindolizin-3-yl)amino]-3-methyl-3-sulfino-butanoic acid, Beta-lactamase OXA-24, TETRAETHYLENE GLYCOL | | Authors: | Romero, A, Santillana, E. | | Deposit date: | 2009-01-24 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design, synthesis, and crystal structures of 6-alkylidene-2'-substituted penicillanic acid sulfones as potent inhibitors of Acinetobacter baumannii OXA-24 carbapenemase.
J.Am.Chem.Soc., 132, 2010
|
|
3FTP
 
 | |
3FT8
 
 | | Structure of HSP90 bound with a noval fragment. | | Descriptor: | (5E,7S)-2-amino-7-(4-fluoro-2-pyridin-3-ylphenyl)-4-methyl-7,8-dihydroquinazolin-5(6H)-one oxime, Heat shock protein HSP 90-alpha | | Authors: | Barker, J.B, Cheng, R.K.Y, Palan, S, Felicetti, B. | | Deposit date: | 2009-01-12 | | Release date: | 2009-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fragment-based identification of Hsp90 inhibitors.
Chemmedchem, 4, 2009
|
|
3FT5
 
 | | Structure of HSP90 bound with a novel fragment | | Descriptor: | 4-methyl-7,8-dihydro-5H-thiopyrano[4,3-d]pyrimidin-2-amine, Heat shock protein HSP 90-alpha | | Authors: | Barker, J.B, Mather, O, Cheng, R.K.Y, Palan, S, Felicetti, B. | | Deposit date: | 2009-01-12 | | Release date: | 2009-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fragment-based Identification of Hsp90 Inhibitors.
Chemmedchem, 4, 2009
|
|
1OJ7
 
 | | STRUCTURAL GENOMICS, UNKNOWN FUNCTION CRYSTAL STRUCTURE OF E. COLI K-12 YQHD | | Descriptor: | 5,6-DIHYDROXY-NADP, BORIC ACID, CHLORIDE ION, ... | | Authors: | Sulzenbacher, G, Perrier, S, Roig-Zamboni, V, Pagot, F, Grisel, S, Salamoni, A, Valencia, C, Bignon, C, Vincentelli, R, Tegoni, M, Cambillau, C. | | Deposit date: | 2003-07-03 | | Release date: | 2004-07-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of E.Coli Alcohol Dehydrogenase Yqhd: Evidence of a Covalently Modified Nadp Coenzyme
J.Mol.Biol., 342, 2004
|
|
1OHG
 
 | | STRUCTURE OF THE DSDNA BACTERIOPHAGE HK97 MATURE EMPTY CAPSID | | Descriptor: | CHLORIDE ION, MAJOR CAPSID PROTEIN, SULFATE ION | | Authors: | Helgstrand, C, Wikoff, W.R, Duda, R.L, Hendrix, R.W, Johnson, J.E, Liljas, L. | | Deposit date: | 2003-05-26 | | Release date: | 2003-12-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | The Refined Structure of a Protein Catenane: The Hk97 Bacteriophage Capsid at 3.44A Resolution
J.Mol.Biol., 334, 2003
|
|
1PEH
 
 | | NMR STRUCTURE OF THE MEMBRANE-BINDING DOMAIN OF CTP PHOSPHOCHOLINE CYTIDYLYLTRANSFERASE, 10 STRUCTURES | | Descriptor: | PEPNH1 | | Authors: | Dunne, S.J, Cornell, R.B, Johnson, J.E, Glover, N.R, Tracey, A.S. | | Deposit date: | 1996-06-10 | | Release date: | 1996-12-07 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Structure of the membrane binding domain of CTP:phosphocholine cytidylyltransferase.
Biochemistry, 35, 1996
|
|
