7KA0
 
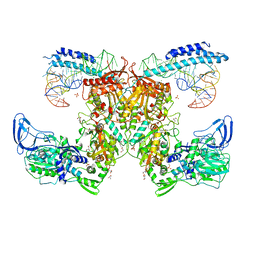 | | Crystal structure of the complex of M. tuberculosis PheRS with cognate precursor tRNA and phenylalanine | | Descriptor: | ACETATE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Chang, C, Michalska, K, Jedrzejczak, R, Wower, J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-09-29 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mycobacterium tuberculosis Phe-tRNA synthetase: structural insights into tRNA recognition and aminoacylation.
Nucleic Acids Res., 49, 2021
|
|
3DHS
 
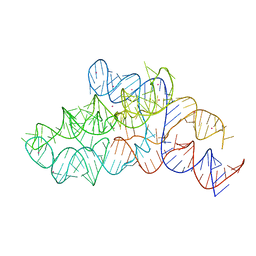 | |
5EBI
 
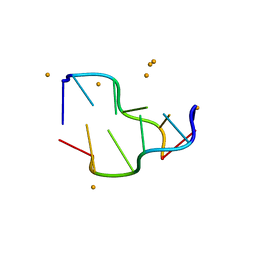 | | Crystal structure of a DNA-RNA chimera in complex with Ba2+ ions: a case of unusual multi-domain twinning | | Descriptor: | BARIUM ION, DNA/RNA (5'-D(*C)-R(P*G)-D(P*C)-R(P*G)-D(P*C)-R(P*G)-3') | | Authors: | Gilski, M, Drozdzal, P, Kierzek, R, Jaskolski, M. | | Deposit date: | 2015-10-19 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Atomic resolution structure of a chimeric DNA-RNA Z-type duplex in complex with Ba(2+) ions: a case of complicated multi-domain twinning.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
6URO
 
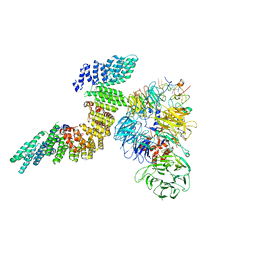 | | Cryo-EM structure of human CPSF160-WDR33-CPSF30-PAS RNA-CstF77 complex | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 1, Cleavage and polyadenylation specificity factor subunit 4, Cleavage stimulation factor subunit 3, ... | | Authors: | Sun, Y, Zhang, Y, Walz, T, Tong, L. | | Deposit date: | 2019-10-23 | | Release date: | 2019-11-27 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Insights into the Human Pre-mRNA 3'-End Processing Machinery.
Mol.Cell, 77, 2020
|
|
7CV1
 
 | |
9DSX
 
 | | Crystal structure of the complex of M. tuberculosis PheRS with cognate precursor tRNA and fragment DDD00107555 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Chang, C, Michalska, K, Forte, B, Baragana, B, Gilbert, I.H, Wower, J, Joachimiak, A. | | Deposit date: | 2024-09-30 | | Release date: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Different chemical scaffolds bind to L-phe site in Mycobacterium tuberculosis Phe-tRNA synthetase.
Eur.J.Med.Chem., 287, 2025
|
|
9DTF
 
 | | Crystal structure of the complex of M. tuberculosis PheRS with cognate precursor tRNA and fragment DDD01008876 | | Descriptor: | (5S)-7-benzyl-1,3,7-triazaspiro[4.4]nonane-2,4-dione, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Chang, C, Michalska, K, Forte, B, Baragana, B, Gilbert, I.H, Wower, J, Joachimiak, A. | | Deposit date: | 2024-09-30 | | Release date: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Different chemical scaffolds bind to L-phe site in Mycobacterium tuberculosis Phe-tRNA synthetase.
Eur.J.Med.Chem., 287, 2025
|
|
9DRT
 
 | | Crystal structure of the complex of M. tuberculosis PheRS with cognate precursor tRNA and fragment DDD00805735 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Chang, C, Michalska, K, Forte, B, Baragana, B, Gilbert, I.H, Wower, J, Joachimiak, A, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-09-26 | | Release date: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Different chemical scaffolds bind to L-phe site in Mycobacterium tuberculosis Phe-tRNA synthetase.
Eur.J.Med.Chem., 287, 2025
|
|
6YMT
 
 | | RNASE 3/1 version1 | | Descriptor: | GLYCEROL, RNase 3/1 version 1 | | Authors: | Fernandez-Millan, P, Prats-Ejarque, G, Vazquez-Monteagudo, S, Boix, E. | | Deposit date: | 2020-04-09 | | Release date: | 2021-07-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Exploring the RNase A scaffold to combine catalytic and antimicrobial activities. Structural characterization of RNase 3/1 chimeras.
Front Mol Biosci, 9, 2022
|
|
6YBC
 
 | | RNASE 3/1 version2 phosphate complex | | Descriptor: | GLYCEROL, PHOSPHATE ION, RNASE 3/1 version2 | | Authors: | Fernandez-Millan, P, Prats-Ejarque, G, Vazquez-Monteagudo, S, Boix, E. | | Deposit date: | 2020-03-16 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Exploring the RNase A scaffold to combine catalytic and antimicrobial activities. Structural characterization of RNase 3/1 chimeras.
Front Mol Biosci, 9, 2022
|
|
8P7D
 
 | | CryoEM structure of METTL6 tRNA SerRS complex in a 1:1:2 stoichiometry | | Descriptor: | MAGNESIUM ION, S-ADENOSYL-L-HOMOCYSTEINE, Serine tRNA, ... | | Authors: | Throll, P, Dolce, L.G, Kowalinski, E. | | Deposit date: | 2023-05-30 | | Release date: | 2024-06-12 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural basis of tRNA recognition by the m 3 C RNA methyltransferase METTL6 in complex with SerRS seryl-tRNA synthetase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8P7B
 
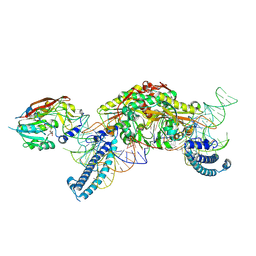 | | CryoEM structure of METTL6 tRNA SerRS complex in a 1:2:2 stoichiometry | | Descriptor: | MAGNESIUM ION, S-ADENOSYL-L-HOMOCYSTEINE, Serine tRNA, ... | | Authors: | Throll, P, Dolce, L.G, Kowalinski, E. | | Deposit date: | 2023-05-30 | | Release date: | 2024-06-12 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.42 Å) | | Cite: | Structural basis of tRNA recognition by the m 3 C RNA methyltransferase METTL6 in complex with SerRS seryl-tRNA synthetase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8P7C
 
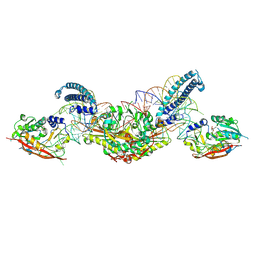 | | CryoEM structure of METTL6 tRNA SerRS complex in a 2:2:2 stoichiometry | | Descriptor: | MAGNESIUM ION, S-ADENOSYL-L-HOMOCYSTEINE, Serine tRNA, ... | | Authors: | Throll, P, Dolce, L.G, Kowalinski, E. | | Deposit date: | 2023-05-30 | | Release date: | 2024-06-12 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of tRNA recognition by the m 3 C RNA methyltransferase METTL6 in complex with SerRS seryl-tRNA synthetase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6VA3
 
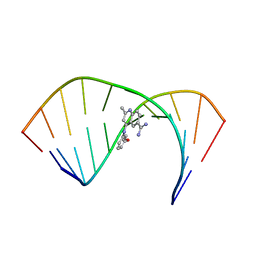 | | Solution Structure of the Tau pre-mRNA Exon 10 Splicing Regulatory Element Bound to MQC | | Descriptor: | 4-[(3-methoxyphenyl)amino]-2-methylquinoline-6-carboximidamide, RNA (5'-R(*CP*AP*CP*AP*CP*GP*UP*CP*GP*G)-3'), RNA (5'-R(*CP*CP*GP*GP*CP*AP*GP*UP*GP*UP*G)-3') | | Authors: | Chen, J.L, Fountain, M.A, Disney, M.D. | | Deposit date: | 2019-12-16 | | Release date: | 2020-05-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Design, Optimization, and Study of Small Molecules That Target Tau Pre-mRNA and Affect Splicing.
J.Am.Chem.Soc., 142, 2020
|
|
6VA4
 
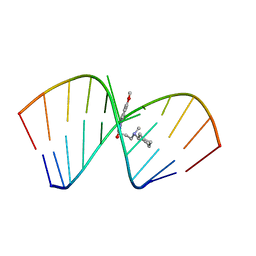 | | Solution Structure of the Tau pre-mRNA Exon 10 Splicing Regulatory Element Bound to MIP | | Descriptor: | N-[3-(8-methoxy-4-oxo-4,5-dihydro-3H-pyrimido[5,4-b]indol-3-yl)propyl]-N-methylcyclohexanaminium, RNA (5'-R(*CP*AP*CP*AP*CP*GP*UP*CP*GP*G)-3'), RNA (5'-R(*CP*CP*GP*GP*CP*AP*GP*UP*GP*UP*G)-3') | | Authors: | Chen, J.L, Fountain, M.A, Disney, M.D. | | Deposit date: | 2019-12-16 | | Release date: | 2020-05-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Design, Optimization, and Study of Small Molecules That Target Tau Pre-mRNA and Affect Splicing.
J.Am.Chem.Soc., 142, 2020
|
|
6VA2
 
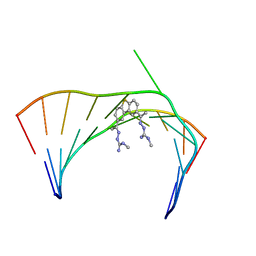 | | Solution Structure of the Tau pre-mRNA Exon 10 Splicing Regulatory Element Bound to MH5 | | Descriptor: | (2E,2'E)-2,2'-{dibenzo[b,d]thiene-2,8-diyldi[(1E)eth-1-yl-1-ylidene]}bis(N-methylhydrazine-1-carboximidamide), RNA (5'-R(*CP*AP*CP*AP*CP*GP*UP*CP*GP*G)-3'), RNA (5'-R(*CP*CP*GP*GP*CP*AP*GP*UP*GP*UP*G)-3') | | Authors: | Chen, J.L, Fountain, M.A, Disney, M.D. | | Deposit date: | 2019-12-16 | | Release date: | 2020-05-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Design, Optimization, and Study of Small Molecules That Target Tau Pre-mRNA and Affect Splicing.
J.Am.Chem.Soc., 142, 2020
|
|
6JKI
 
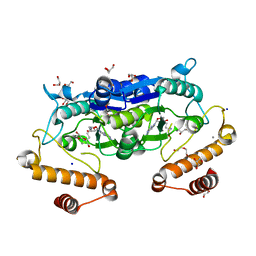 | | Crystal structure and catalytic mechanism of the essential m1G37 tRNA methyltransferase TrmD from Pseudomonas aeruginosa | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Jaroensuk, J, Liew, C.W, Atichartpongkul, S, Chionh, Y.H, Wong, Y.H, Zhong, W.H, McBee, M.E, Thongdee, N, Prestwich, E.G, DeMott, M.S, Mongkolsuk, S, Dedon, P.C, Lescar, J, Fuangthong, M. | | Deposit date: | 2019-03-01 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure and catalytic mechanism of the essential m1G37 tRNA methyltransferase TrmD fromPseudomonas aeruginosa.
Rna, 25, 2019
|
|
3TS1
 
 | |
5WYQ
 
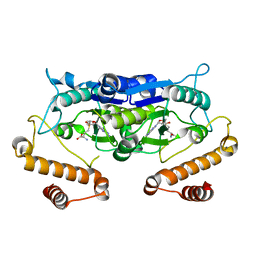 | | Crystal Structure and catalytic mechanism of the essential m1G37 tRNA methyltransferase TrmD from Pseudomonas aeruginosa | | Descriptor: | S-ADENOSYLMETHIONINE, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Jaroensuk, J, Liew, C.W, Atichartpongkul, S, Chionh, Y.H, Wong, Y.H, Zhong, W, McBee, M.E, Maenpuen, S, Prestwich, E.G, DeMott, M.S, Mongkolsuk, S, Dedon, P.C, Lescar, J, Fuangthong, M, Chaiyen, P. | | Deposit date: | 2017-01-15 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure and catalytic mechanism of the essential m1G37 tRNA methyltransferase TrmD fromPseudomonas aeruginosa.
Rna, 2019
|
|
7YNW
 
 | |
5WYR
 
 | | Crystal structure and catalytic mechanism of the essential m1G37 tRNA methyltransferase TrmD from Pseudomonas aeruginosa | | Descriptor: | SINEFUNGIN, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Jaroensuk, J, Liew, C.W, Atichartpongkul, S, Chionh, Y.H, Wong, Y.H, Zhong, W.H, McBee, M.E, Thongdee, N, Prestwich, E.G, DeMott, M.S, Mongkolsuk, S, Dedon, P.C, Lescar, J, Fuangthong, M. | | Deposit date: | 2017-01-15 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure and catalytic mechanism of the essential m1G37 tRNA methyltransferase TrmD fromPseudomonas aeruginosa.
Rna, 2019
|
|
7PMM
 
 | |
7O4O
 
 | | Structure of Staphylococcus aureus m1A22-tRNA methyltransferase in complex with S-adenosylhomocysteine | | Descriptor: | GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, tRNA (Adenine(22)-N(1))-methyltransferase | | Authors: | Gloster, T.M, Czekster, C.M, da Silva, R.G. | | Deposit date: | 2021-04-06 | | Release date: | 2022-04-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structure, dynamics, and molecular inhibition of the Staphylococcus aureus m 1 A22-tRNA methyltransferase TrmK.
J.Biol.Chem., 298, 2022
|
|
7O4N
 
 | | Structure of Staphylococcus aureus m1A22-tRNA methyltransferase in complex with S-adenosylmethionine | | Descriptor: | GLYCEROL, S-ADENOSYLMETHIONINE, tRNA (Adenine(22)-N(1))-methyltransferase | | Authors: | Gloster, T.M, Czekster, C.M, da Silva, R.G. | | Deposit date: | 2021-04-06 | | Release date: | 2022-04-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure, dynamics, and molecular inhibition of the Staphylococcus aureus m 1 A22-tRNA methyltransferase TrmK.
J.Biol.Chem., 298, 2022
|
|
7O4M
 
 | | Structure of Staphylococcus aureus m1A22-tRNA methyltransferase | | Descriptor: | CITRIC ACID, GLYCEROL, tRNA (Adenine(22)-N(1))-methyltransferase | | Authors: | Gloster, T.M, Czekster, C.M, da Silva, R.G. | | Deposit date: | 2021-04-06 | | Release date: | 2022-04-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure, dynamics, and molecular inhibition of the Staphylococcus aureus m 1 A22-tRNA methyltransferase TrmK.
J.Biol.Chem., 298, 2022
|
|
