6SCS
 
 | |
5IKX
 
 | |
6YMY
 
 | | Cytochrome c oxidase from Saccharomyces cerevisiae | | Descriptor: | (2R,5S,11R,14R)-5,8,11-trihydroxy-2-(nonanoyloxy)-5,11-dioxido-16-oxo-14-[(propanoyloxy)methyl]-4,6,10,12,15-pentaoxa-5,11-diphosphanonadec-1-yl undecanoate, 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, COPPER (II) ION, ... | | Authors: | Berndtsson, J, Rathore, S, Ott, M. | | Deposit date: | 2020-04-10 | | Release date: | 2020-09-09 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Respiratory supercomplexes enhance electron transport by decreasing cytochrome c diffusion distance.
Embo Rep., 21, 2020
|
|
5II8
 
 | | Orthorhombic crystal structure of red abalone lysin at 0.99 A resolution | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Egg-lysin, SULFATE ION | | Authors: | Sadat Al-Hosseini, H, Raj, I, Nishimura, K, De Sanctis, D, Jovine, L. | | Deposit date: | 2016-03-01 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Structural Basis of Egg Coat-Sperm Recognition at Fertilization.
Cell, 169, 2017
|
|
7U8G
 
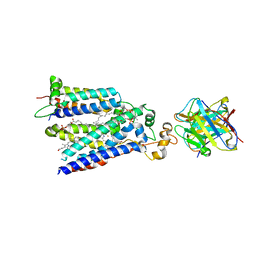 | | Cryo-EM structure of the core human NADPH oxidase NOX2 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 7G5 - heavy chain, ... | | Authors: | Noreng, S, Ota, N, Sun, Y, Masureel, M, Payandeh, J, Yi, T, Koerber, J.T. | | Deposit date: | 2022-03-08 | | Release date: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the core human NADPH oxidase NOX2.
Nat Commun, 13, 2022
|
|
4RDK
 
 | | Crystal structure of Norovirus Boxer P domain in complex with Lewis b tetrasaccharide | | Descriptor: | Capsid, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Hao, N, Chen, Y, Xia, M, Liu, W, Tan, M, Jiang, X, Li, X. | | Deposit date: | 2014-09-19 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.629 Å) | | Cite: | Crystal structures of GI.8 Boxer virus P dimers in complex with HBGAs, a novel evolutionary path selected by the Lewis epitope.
Protein Cell, 6, 2015
|
|
6AU8
 
 | |
6SFW
 
 | |
6Y5D
 
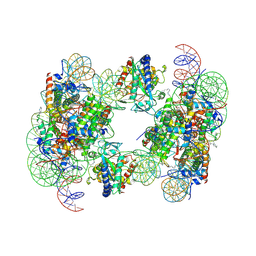 | | Structure of human cGAS (K394E) bound to the nucleosome | | Descriptor: | Cyclic GMP-AMP synthase, DNA (153-MER), Histone H2A type 2-A, ... | | Authors: | Pathare, G.R, Cavadini, S, Kempf, G, Thoma, N.H. | | Deposit date: | 2020-02-25 | | Release date: | 2020-09-23 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural mechanism of cGAS inhibition by the nucleosome.
Nature, 587, 2020
|
|
5MFU
 
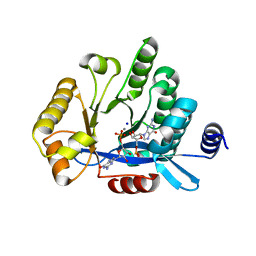 | | PA3825-EAL Mn-pGpG Structure | | Descriptor: | Diguanylate phosphodiesterase, GUANOSINE-5'-MONOPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Horrell, S, Bellini, D, Strange, R, Wagner, A, Walsh, M. | | Deposit date: | 2016-11-18 | | Release date: | 2017-03-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Dimerisation induced formation of the active site and the identification of three metal sites in EAL-phosphodiesterases.
Sci Rep, 7, 2017
|
|
6B28
 
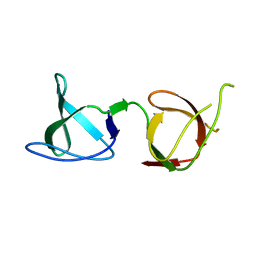 | |
6SFX
 
 | |
5MR3
 
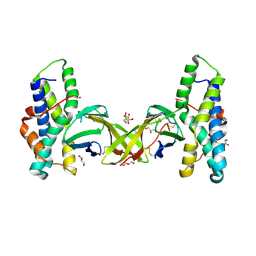 | | Crystal structure of red abalone egg VERL repeat 2 with linker in complex with sperm lysin at 1.8 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Egg-lysin, ... | | Authors: | Nishimura, K, Raj, I, Sadat Al-Hosseini, H, De Sanctis, D, Jovine, L. | | Deposit date: | 2016-12-21 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Egg Coat-Sperm Recognition at Fertilization.
Cell, 169, 2017
|
|
6SS2
 
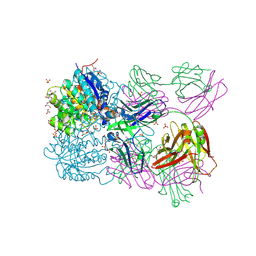 | | Structure of arginase-2 in complex with the inhibitory human antigen-binding fragment Fab C0021158 | | Descriptor: | Arginase-2, mitochondrial, Fab C0021158 heavy chain (IgG1), ... | | Authors: | Burschowsky, D, Addyman, A, Fiedler, S, Groves, M, Haynes, S, Seewooruthun, C, Carr, M. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional characterization of C0021158, a high-affinity monoclonal antibody that inhibits Arginase 2 function via a novel non-competitive mechanism of action.
Mabs, 12
|
|
5IZJ
 
 | | Complex of PKA with the bisubstrate protein kinase inhibitor ARC-1411 | | Descriptor: | 4-(piperazin-1-yl)-7H-pyrrolo[2,3-d]pyrimidine, 47P-AZ1-DAR-DAR, 47P-AZ1-DAR-DAR-DAR, ... | | Authors: | Pflug, A, Enkvist, E, Uri, A, Engh, R.A. | | Deposit date: | 2016-03-25 | | Release date: | 2016-07-20 | | Last modified: | 2019-06-19 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Bifunctional Ligands for Inhibition of Tight-Binding Protein-Protein Interactions.
Bioconjug.Chem., 27, 2016
|
|
5MKG
 
 | | PA3825-EAL Ca-CdG Structure | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CALCIUM ION, Diguanylate phosphodiesterase | | Authors: | Horrell, S, Bellini, D, Strange, R, Wagner, A, Walsh, M. | | Deposit date: | 2016-12-04 | | Release date: | 2016-12-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Dimerisation induced formation of the active site and the identification of three metal sites in EAL-phosphodiesterases.
Sci Rep, 7, 2017
|
|
6YGB
 
 | | Crystal structure of the NatC complex bound to CoA | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, COENZYME A, ... | | Authors: | Grunwald, S, Hopf, L, Bock-Bierbaum, T, Lally, C.C, Spahn, C.M.T, Daumke, O. | | Deposit date: | 2020-03-27 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.451 Å) | | Cite: | Divergent architecture of the heterotrimeric NatC complex explains N-terminal acetylation of cognate substrates.
Nat Commun, 11, 2020
|
|
6YGI
 
 | |
5IZF
 
 | | Complex of PKA with the bisubstrate protein kinase inhibitor ARC-1408 | | Descriptor: | 6J9-ZEU-DAR-ACA-DAR-NH2, SULFATE ION, cAMP-dependent protein kinase catalytic subunit alpha | | Authors: | Pflug, A, Enkvist, E, Uri, A, Engh, R.A. | | Deposit date: | 2016-03-25 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Bifunctional Ligands for Inhibition of Tight-Binding Protein-Protein Interactions.
Bioconjug.Chem., 27, 2016
|
|
6MGT
 
 | | Crystal structure of alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde Decarboxylase Mutant H110A | | Descriptor: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, COBALT (II) ION | | Authors: | Yang, Y, Daivs, I, Matsui, T, Rubalcava, I, Liu, A. | | Deposit date: | 2018-09-14 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Quaternary structure of alpha-amino-beta-carboxymuconate-ε-semialdehyde decarboxylase (ACMSD) controls its activity.
J.Biol.Chem., 294, 2019
|
|
6YPU
 
 | | Acinetobacter baumannii ribosome-amikacin complex - 30S subunit body | | Descriptor: | (2S)-N-[(1R,2S,3S,4R,5S)-4-[(2R,3R,4S,5S,6R)-6-(aminomethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-5-azanyl-2-[(2S,3R,4S,5S ,6R)-4-azanyl-6-(hydroxymethyl)-3,5-bis(oxidanyl)oxan-2-yl]oxy-3-oxidanyl-cyclohexyl]-4-azanyl-2-oxidanyl-butanamide, 16S ribosomal RNA, 30S ribosomal protein S11, ... | | Authors: | Nicholson, D, Edwards, T.A, O'Neill, A.J, Ranson, N.A. | | Deposit date: | 2020-04-16 | | Release date: | 2020-09-16 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of the 70S Ribosome from the Human Pathogen Acinetobacter baumannii in Complex with Clinically Relevant Antibiotics.
Structure, 28, 2020
|
|
7WSW
 
 | | Cryo-EM structure of the Potassium channel AKT1 from Arabidopsis thaliana | | Descriptor: | PHOSPHATIDYLETHANOLAMINE, POTASSIUM ION, Potassium channel AKT1 | | Authors: | Yang, G.H, Lu, Y.M, Zhang, Y.M, Jia, Y.T, Li, X.M, Lei, J.L. | | Deposit date: | 2022-02-02 | | Release date: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for the activity regulation of a potassium channel AKT1 from Arabidopsis.
Nat Commun, 13, 2022
|
|
7BO9
 
 | |
6SS0
 
 | | Structure of the arginase-2-inhibitory human antigen-binding fragment Fab C0021181 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Fab C0021181 heavy chain (IgG1), ... | | Authors: | Burschowsky, D, Addyman, A, Fiedler, S, Groves, M, Haynes, S, Seewooruthun, C, Carr, M. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and functional characterization of C0021158, a high-affinity monoclonal antibody that inhibits Arginase 2 function via a novel non-competitive mechanism of action.
Mabs, 12
|
|
5MFQ
 
 | | Crystal structure of the GluK1 ligand-binding domain in complex with kainate and BPAM-344 at 1.90 A resolution | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, 4-cyclopropyl-7-fluoro-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-dioxide, CHLORIDE ION, ... | | Authors: | Larsen, A.P, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2016-11-18 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification and Structure-Function Study of Positive Allosteric Modulators of Kainate Receptors.
Mol. Pharmacol., 91, 2017
|
|
