3MOK
 
 | | Structure of Apo HasAp from Pseudomonas aeruginosa to 1.55A Resolution | | Descriptor: | Heme acquisition protein HasAp, PHOSPHATE ION, SODIUM ION | | Authors: | Lovell, S, Battaile, K.P, Jepkorir, G, Rodriguez, J.C, Rui, H, Im, W, Alontaga, A.Y, Yukl, E, Moenne-Loccoz, P, Rivera, M. | | Deposit date: | 2010-04-22 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural, NMR Spectroscopic, and Computational Investigation of Hemin Loading in the Hemophore HasAp from Pseudomonas aeruginosa.
J.Am.Chem.Soc., 132, 2010
|
|
3V56
 
 | | Re-refinement of PDB entry 1OSG - Complex between BAFF and a BR3 derived peptide presented in a beta-hairpin scaffold - reveals an additonal copy of the peptide. | | Descriptor: | BR3 derived peptive, SULFATE ION, Tumor necrosis factor ligand superfamily member 13B | | Authors: | Smart, O.S, Womack, T.O, Flensburg, C, Keller, P, Sharff, A, Paciorek, W, Vonrhein, C, Bricogne, G. | | Deposit date: | 2011-12-16 | | Release date: | 2012-03-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Exploiting structure similarity in refinement: automated NCS and target-structure restraints in BUSTER.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4OEX
 
 | | Crystal structure of the PDE5A1 catalytic domain in complex with novel inhibitors | | Descriptor: | 6-ethyl-2-{5-[(4-methylpiperazin-1-yl)sulfonyl]-2-propoxyphenyl}pyrimidin-4(3H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Chen, T.T, Ren, J, Xu, Y.C. | | Deposit date: | 2014-01-14 | | Release date: | 2015-04-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Thermodynamic and structural characterization of halogen bonding in protein-ligand interactions: a case study of PDE5 and its inhibitors.
J.Med.Chem., 57, 2014
|
|
4RMH
 
 | | Human Sirt2 in complex with SirReal2 and Ac-Lys-H3 peptide | | Descriptor: | 2-[(4,6-dimethylpyrimidin-2-yl)sulfanyl]-N-[5-(naphthalen-1-ylmethyl)-1,3-thiazol-2-yl]acetamide, Ac-Lys-H3 peptide, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Rumpf, T, Schiedel, M, Karaman, B, Roessler, C, North, B.J, Lehotzky, A, Olah, J, Ladwein, K.I, Schmidtkunz, K, Gajer, M, Pannek, M, Steegborn, C, Sinclair, D.A, Gerhardt, S, Ovadi, J, Schutkowski, M, Sippl, W, Einsle, O, Jung, M. | | Deposit date: | 2014-10-21 | | Release date: | 2015-02-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Selective Sirt2 inhibition by ligand-induced rearrangement of the active site.
Nat Commun, 6, 2015
|
|
4RMJ
 
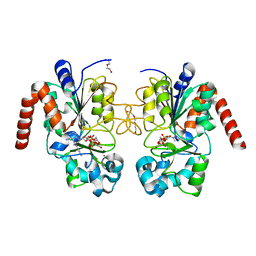 | | Human Sirt2 in complex with ADP ribose and nicotinamide | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Rumpf, T, Schiedel, M, Karaman, B, Roessler, C, North, B.J, Lehotzky, A, Olah, J, Ladwein, K.I, Schmidtkunz, K, Gajer, M, Pannek, M, Steegborn, C, Sinclair, D.A, Gerhardt, S, Ovadi, J, Schutkowski, M, Sippl, W, Einsle, O, Jung, M. | | Deposit date: | 2014-10-21 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Selective Sirt2 inhibition by ligand-induced rearrangement of the active site.
Nat Commun, 6, 2015
|
|
4R9B
 
 | |
1P7K
 
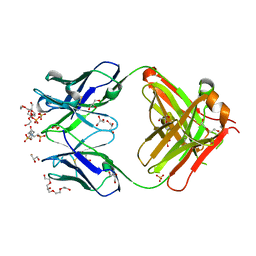 | | Crystal structure of an anti-ssDNA antigen-binding fragment (Fab) bound to 4-(2-Hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Schuermann, J.P, Henzl, M.T, Deutscher, S.L, Tanner, J.J. | | Deposit date: | 2003-05-02 | | Release date: | 2004-05-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of an anti-DNA fab complexed with a non-DNA ligand provides insights into cross-reactivity and molecular mimicry.
Proteins, 57, 2004
|
|
4RL7
 
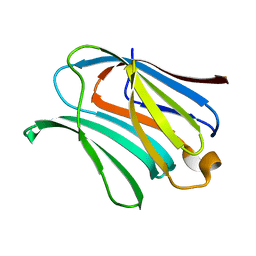 | |
4B5N
 
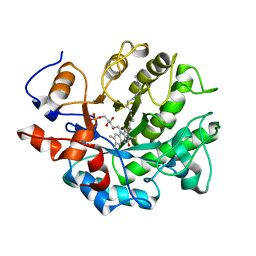 | | Crystal structure of oxidized Shewanella Yellow Enzyme 4 (SYE4) | | Descriptor: | FLAVIN MONONUCLEOTIDE, OXIDOREDUCTASE, FMN-BINDING, ... | | Authors: | Elegheert, J, Brige, A, Savvides, S.N. | | Deposit date: | 2012-08-07 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural dissection of Shewanella oneidensis old yellow enzyme 4 bound to a Meisenheimer complex and (nitro)phenolic ligands.
FEBS Lett., 591, 2017
|
|
7CR0
 
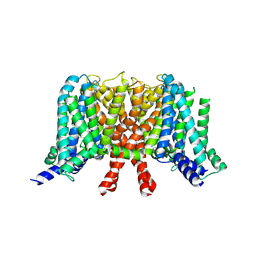 | | human KCNQ2 in apo state | | Descriptor: | Potassium voltage-gated channel subfamily KQT member 2 | | Authors: | Li, X, Lv, D, Wang, J, Ye, S, Guo, J. | | Deposit date: | 2020-08-12 | | Release date: | 2020-09-16 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular basis for ligand activation of the human KCNQ2 channel.
Cell Res., 31, 2021
|
|
7CR3
 
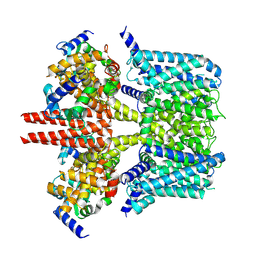 | | human KCNQ2-CaM in apo state | | Descriptor: | Calmodulin-3, Potassium voltage-gated channel subfamily KQT member 2 | | Authors: | Li, X, Lv, D, Wang, J, Ye, S, Guo, J. | | Deposit date: | 2020-08-12 | | Release date: | 2020-09-16 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular basis for ligand activation of the human KCNQ2 channel.
Cell Res., 31, 2021
|
|
3WFE
 
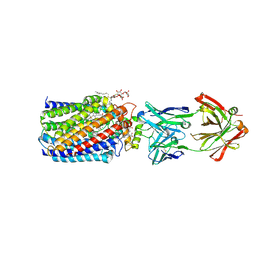 | | Reduced and cyanide-bound cytochrome c-dependent nitric oxide reductase (cNOR) from Pseudomonas aeruginosa in complex with antibody fragment | | Descriptor: | CALCIUM ION, CYANIDE ION, FE (III) ION, ... | | Authors: | Sato, N, Ishii, S, Hino, T, Sugimoto, H, Fukumori, Y, Shiro, Y, Tosha, T. | | Deposit date: | 2013-07-18 | | Release date: | 2014-05-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structures of reduced and ligand-bound nitric oxide reductase provide insights into functional differences in respiratory enzymes.
Proteins, 82, 2014
|
|
7CR4
 
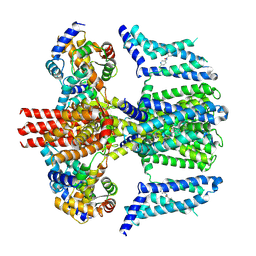 | | human KCNQ2-CaM in complex with ztz240 | | Descriptor: | Calmodulin-3, N-(6-chloranylpyridin-3-yl)-4-fluoranyl-benzamide, Potassium voltage-gated channel subfamily KQT member 2 | | Authors: | Li, X, Lv, D, Wang, J, Ye, S, Guo, J. | | Deposit date: | 2020-08-12 | | Release date: | 2020-09-16 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Molecular basis for ligand activation of the human KCNQ2 channel.
Cell Res., 31, 2021
|
|
1MFL
 
 | |
4R9D
 
 | |
6BN0
 
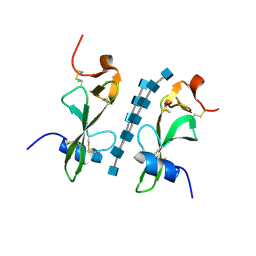 | | Avirulence protein 4 (Avr4) from Cladosporium fulvum bound to the hexasaccharide of chitin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Race-specific elicitor A4 | | Authors: | Hurlburt, N.K, Fisher, A.J. | | Deposit date: | 2017-11-15 | | Release date: | 2018-08-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the Cladosporium fulvum Avr4 effector in complex with (GlcNAc)6 reveals the ligand-binding mechanism and uncouples its intrinsic function from recognition by the Cf-4 resistance protein.
PLoS Pathog., 14, 2018
|
|
3WFB
 
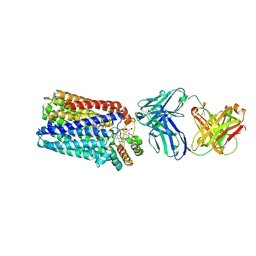 | | Reduced cytochrome c-dependent nitric oxide reductase (cNOR) from Pseudomonas aeruginosa in complex with antibody fragment | | Descriptor: | CALCIUM ION, CHLORIDE ION, FE (III) ION, ... | | Authors: | Sato, N, Ishii, S, Hino, T, Sugimoto, H, Fukumori, Y, Shiro, Y, Tosha, T. | | Deposit date: | 2013-07-18 | | Release date: | 2014-05-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of reduced and ligand-bound nitric oxide reductase provide insights into functional differences in respiratory enzymes.
Proteins, 82, 2014
|
|
3WFC
 
 | | Reduced and carbonmonoxide-bound cytochrome c-dependent nitric oxide reductase (cNOR) from Pseudomonas aeruginosa in complex with antibody fragment | | Descriptor: | CALCIUM ION, CARBON MONOXIDE, FE (III) ION, ... | | Authors: | Sato, N, Ishii, S, Hino, T, Sugimoto, H, Fukumori, Y, Shiro, Y, Tosha, T. | | Deposit date: | 2013-07-18 | | Release date: | 2014-05-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of reduced and ligand-bound nitric oxide reductase provide insights into functional differences in respiratory enzymes.
Proteins, 82, 2014
|
|
3WFD
 
 | | Reduced and acetaldoxime-bound cytochrome c-dependent nitric oxide reductase (cNOR) from Pseudomonas aeruginosa in complex with antibody fragment | | Descriptor: | (1E)-N-hydroxyethanimine, CALCIUM ION, FE (III) ION, ... | | Authors: | Sato, N, Ishii, S, Hino, T, Sugimoto, H, Fukumori, Y, Shiro, Y, Tosha, T. | | Deposit date: | 2013-07-18 | | Release date: | 2014-05-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of reduced and ligand-bound nitric oxide reductase provide insights into functional differences in respiratory enzymes.
Proteins, 82, 2014
|
|
6QKN
 
 | | Structure of the azide-inhibited form of cytochrome c peroxidase from obligate human pathogenic bacterium Neisseria gonorrhoeae | | Descriptor: | AZIDE ION, CALCIUM ION, Cytochrome-c peroxidase, ... | | Authors: | Carvalho, A.L, Romao, M.J, Pauleta, S, Nobrega, C. | | Deposit date: | 2019-01-29 | | Release date: | 2020-02-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the mixed-valence, active form, of cytochrome c peroxidase from obligate human pathogenic bacterium Neisseria gonorrhoeae
To Be Published
|
|
1I8P
 
 | |
3WYY
 
 | | CRYSTAL STRUCTURE OF HUMAN MPS1 CATALYTIC DOMAIN IN COMPLEX WITH (E)-3-(4-((6-(((3s,5s,7s)-adamantan-1-yl)amino)-4-amino-5-cyanopyridin-2-yl)amino)-2-(cyanomethoxy)phenyl)-N-(2-methoxyethyl)acrylamide | | Descriptor: | (2E)-3-[4-({4-amino-5-cyano-6-[(3s,5s,7s)-tricyclo[3.3.1.1~3,7~]dec-1-ylamino]pyridin-2-yl}amino)-2-(cyanomethoxy)phenyl]-N-(2-methoxyethyl)prop-2-enamide, Dual specificity protein kinase TTK | | Authors: | Kusakabe, K, Ide, N, Daigo, Y, Itoh, T, Yamamoto, T, Kojima, E, Mitsuoka, Y, Tadano, G, Tagashira, S, Higashino, K, Okano, Y, Sato, Y, Inoue, M, Iguchi, M, Kanazawa, T, Ishioka, Y, Dohi, K, Kido, Y, Sakamoto, S, Ando, S, Maeda, M, Higaki, M, Yoshizawa, H, Murai, H, Nakamura, Y. | | Deposit date: | 2014-09-10 | | Release date: | 2015-04-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | A unique hinge binder of extremely selective aminopyridine-based Mps1 (TTK) kinase inhibitors with cellular activity.
Bioorg.Med.Chem., 23, 2015
|
|
6FKP
 
 | | Crystal structure of BAZ2A PHD zinc finger in complex with H3 10-mer AA mutant peptide | | Descriptor: | ALA-ARG-THR-ALA-ALA-THR-ALA-ARG, Bromodomain adjacent to zinc finger domain protein 2A, GLYCEROL, ... | | Authors: | Amato, A, Lucas, X, Bortoluzzi, A, Wright, D, Ciulli, A. | | Deposit date: | 2018-01-24 | | Release date: | 2018-03-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Targeting Ligandable Pockets on Plant Homeodomain (PHD) Zinc Finger Domains by a Fragment-Based Approach.
ACS Chem. Biol., 13, 2018
|
|
6IAL
 
 | | Porcine E.coli heat-labile enterotoxin B-pentamer in complex with Lacto-N-neohexaose | | Descriptor: | CALCIUM ION, GLYCEROL, Heat-labile enterotoxin B chain, ... | | Authors: | Heim, J.B, Heggelund, J.E, Krengel, U. | | Deposit date: | 2018-11-27 | | Release date: | 2019-02-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Specificity ofEscherichia coliHeat-Labile Enterotoxin Investigated by Single-Site Mutagenesis and Crystallography.
Int J Mol Sci, 20, 2019
|
|
3WYX
 
 | | CRYSTAL STRUCTURE OF HUMAN MPS1 CATALYTIC DOMAIN IN COMPLEX WITH 6-((3-(cyanomethoxy)-4-(1-methyl-1H-pyrazol-4-yl)phenyl)amino)-2-(cyclohexylamino)nicotinonitrile | | Descriptor: | 6-{[3-(cyanomethoxy)-4-(1-methyl-1H-pyrazol-4-yl)phenyl]amino}-2-(cyclohexylamino)pyridine-3-carbonitrile, Dual specificity protein kinase TTK, IODIDE ION | | Authors: | Kusakabe, K, Ide, N, Daigo, Y, Itoh, T, Yamamoto, T, Kojima, E, Mitsuoka, Y, Tadano, G, Tagashira, S, Higashino, K, Okano, Y, Sato, Y, Inoue, M, Iguchi, M, Kanazawa, T, Ishioka, Y, Dohi, K, Kido, Y, Sakamoto, S, Ando, S, Maeda, M, Higaki, M, Yoshizawa, H, Mura, H, Nakamura, Y. | | Deposit date: | 2014-09-09 | | Release date: | 2015-04-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A unique hinge binder of extremely selective aminopyridine-based Mps1 (TTK) kinase inhibitors with cellular activity.
Bioorg.Med.Chem., 23, 2015
|
|
