6X74
 
 | | Rev1 Mg2+-facilitated Product Complex with no monophosphates | | Descriptor: | CHLORIDE ION, DNA (5'-D(*GP*GP*GP*GP*TP*GP*TP*GP*GP*TP*AP*GP*C*)-3'), DNA (5'-D(P*AP*TP*CP*GP*CP*TP*AP*CP*CP*AP*CP*AP*CP*CP*CP*C)-3'), ... | | Authors: | Weaver, T.M, Freudenthal, B.D. | | Deposit date: | 2020-05-29 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Visualizing Rev1 catalyze protein-template DNA synthesis.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6X7D
 
 | |
6L94
 
 | | The structure of the dioxygenase ABH1 from mouse | | Descriptor: | FE (II) ION, Nucleic acid dioxygenase ALKBH1 | | Authors: | Xie, W, Wang, C, Li, H, Zhang, Y. | | Deposit date: | 2019-11-08 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.10012341 Å) | | Cite: | ALKBH1 promotes lung cancer by regulating m6A RNA demethylation.
Biochem Pharmacol, 189, 2021
|
|
1EU1
 
 | | THE CRYSTAL STRUCTURE OF RHODOBACTER SPHAEROIDES DIMETHYLSULFOXIDE REDUCTASE REVEALS TWO DISTINCT MOLYBDENUM COORDINATION ENVIRONMENTS. | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CADMIUM ION, ... | | Authors: | Li, H.K, Temple, K, Rajagopalan, K.V, Schindelin, H. | | Deposit date: | 2000-04-13 | | Release date: | 2000-08-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The 1.3 A Crystal Structure of Rhodobacter sphaeroides Dimethylsulfoxide Reductase Reveals Two Distinct Molybdenum Coordination Environments
J.Am.Chem.Soc., 122, 2000
|
|
6KUC
 
 | | Crystal structure of Plasmodium falciparum histo-aspartic protease (HAP) zymogen (Form 2) | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, HAP protein | | Authors: | Rathore, I, Mishra, V, Bhaumik, P. | | Deposit date: | 2019-08-31 | | Release date: | 2020-05-27 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Activation mechanism of plasmepsins, pepsin-like aspartic proteases from Plasmodium, follows a unique trans-activation pathway.
Febs J., 288, 2021
|
|
1EVU
 
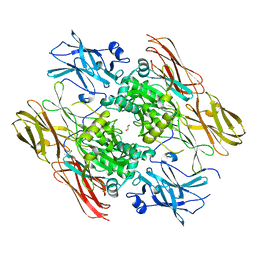 | | HUMAN FACTOR XIII WITH CALCIUM BOUND IN THE ION SITE | | Descriptor: | CALCIUM ION, COAGULATION FACTOR XIII, S-1,2-PROPANEDIOL | | Authors: | Garzon, R.J, Pratt, K.P, Bishop, P.D, Le Trong, I, Stenkamp, R.E, Teller, D.C. | | Deposit date: | 2000-04-20 | | Release date: | 2000-05-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Tryptophan 279 is Essential for the Transglutaminase Activity of Coagulation Factor XIII: Functional and Structural Characterization
To Be Published
|
|
1EP6
 
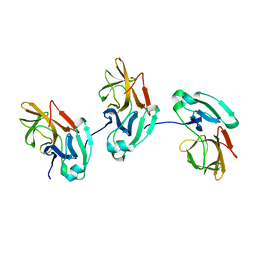 | |
6KOR
 
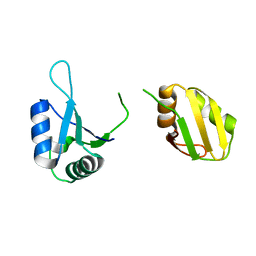 | | Crystal structure of the RRM domain of SYNCRIP | | Descriptor: | Heterogeneous nuclear ribonucleoprotein Q | | Authors: | Chen, Y, Chan, J, Chen, W, Jobichen, C. | | Deposit date: | 2019-08-12 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | SYNCRIP, a new player in pri-let-7a processing.
Rna, 26, 2020
|
|
6XGX
 
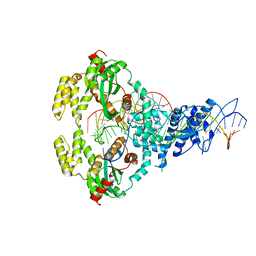 | | ISCth4 transposase, strand transfer complex 1, STC1 | | Descriptor: | DNA (21-MER), DNA (25-MER), DNA (47-MER), ... | | Authors: | Kosek, D, Dyda, F. | | Deposit date: | 2020-06-18 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structures of ISCth4 transpososomes reveal the role of asymmetry in copy-out/paste-in DNA transposition.
Embo J., 40, 2021
|
|
6XNC
 
 | |
6XQS
 
 | | Room-temperature X-ray Crystal structure of SARS-CoV-2 main protease in complex with Telaprevir | | Descriptor: | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide, 3C-like proteinase | | Authors: | Kneller, D.W, Kovalevsky, A, Coates, L. | | Deposit date: | 2020-07-10 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Malleability of the SARS-CoV-2 3CL M pro Active-Site Cavity Facilitates Binding of Clinical Antivirals.
Structure, 28, 2020
|
|
1EVX
 
 | | APO CRYSTAL STRUCTURE OF THE HOMING ENDONUCLEASE, I-PPOI | | Descriptor: | INTRON-ENCODED HOMING ENDONUCLEASE I-PPOI, SULFATE ION, ZINC ION | | Authors: | Galburt, E.A, Jurica, M.S, Chevalier, B.S, Erho, D, Stoddard, B.L. | | Deposit date: | 2000-04-20 | | Release date: | 2000-08-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational changes and cleavage by the homing endonuclease I-PpoI: a critical role for a leucine residue in the active site.
J.Mol.Biol., 300, 2000
|
|
1EI4
 
 | | B-DNA DODECAMER CGCGAAT(TLC)CGCG WITH INCORPORATED [3.3.0]BICYCLO-ARABINO-THYMINE-5'-PHOSPHATE | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*(TLC)P*(TLC)P*CP*GP*CP*G)-3'), MAGNESIUM ION, SPERMINE | | Authors: | Minasov, G, Teplova, M, Nielsen, P, Wengel, J, Egli, M. | | Deposit date: | 2000-02-24 | | Release date: | 2000-03-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structural basis of cleavage by RNase H of hybrids of arabinonucleic acids and RNA.
Biochemistry, 39, 2000
|
|
6XQU
 
 | |
1EX0
 
 | | HUMAN FACTOR XIII, MUTANT W279F ZYMOGEN | | Descriptor: | CALCIUM ION, COAGULATION FACTOR XIII A CHAIN, PHOSPHATE ION, ... | | Authors: | Garzon, R.J, Pratt, K.P, Bishop, P.D, Le Trong, I, Stenkamp, R.E, Teller, D.C. | | Deposit date: | 2000-04-28 | | Release date: | 2003-12-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Tryptophan 279 is Essential for the Transglutaminase Activity of Coagulation Factor XIII: Functional and Structural Characterization
To be Published
|
|
1EXZ
 
 | | STRUCTURE OF STEM CELL FACTOR | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, SAMARIUM (III) ION, ... | | Authors: | Zhang, Z, Zhang, R, Joachimiak, A, Schlessinger, J, Kong, X. | | Deposit date: | 2000-05-05 | | Release date: | 2000-07-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of human stem cell factor: implication for stem cell factor receptor dimerization and activation.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
6KUD
 
 | | Crystal structure of Plasmodium falciparum histo-aspartic protease (HAP) zymogen (Form 3) | | Descriptor: | GLYCEROL, HAP protein | | Authors: | Rathore, I, Mishra, V, Bhaumik, P. | | Deposit date: | 2019-08-31 | | Release date: | 2020-05-27 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Activation mechanism of plasmepsins, pepsin-like aspartic proteases from Plasmodium, follows a unique trans-activation pathway.
Febs J., 288, 2021
|
|
6KUB
 
 | | Crystal structure of Plasmodium falciparum histo-aspartic protease (HAP) zymogen (Form 1) | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, HAP protein | | Authors: | Rathore, I, Mishra, V, Bhaumik, P. | | Deposit date: | 2019-08-31 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Activation mechanism of plasmepsins, pepsin-like aspartic proteases from Plasmodium, follows a unique trans-activation pathway.
Febs J., 288, 2021
|
|
1ENA
 
 | |
6XHU
 
 | |
1E9V
 
 | | XENON BOUND IN HYDROPHOBIC CHANNEL OF HYBRID CLUSTER PROTEIN FROM DESULFOVIBRIO VULGARIS | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ... | | Authors: | Cooper, S.J, Bailey, S, Rizkallah, P.J, Lindley, P.F. | | Deposit date: | 2000-10-27 | | Release date: | 2001-10-25 | | Last modified: | 2020-12-23 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Ferricyanide Soaked Hybrid Cluster Protein at 1.2A and Xenon Mapping of the Hydrophobic Cavity at 1.8A
To be Published
|
|
6X71
 
 | |
6XS3
 
 | |
6KZI
 
 | | Crystal structure of Ser/Thr kinase Pim1 in complex with thioridazine derivatives | | Descriptor: | 4-(2-chloro-10H-phenoxazin-10-yl)-N,N-diethylbutan-1-amine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Wan, X, Li, W, Xie, Y, Huang, N. | | Deposit date: | 2019-09-24 | | Release date: | 2020-03-04 | | Last modified: | 2021-03-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
1EKJ
 
 | |
