2JGW
 
 | | Structure of CCP module 7 of complement factor H - The AMD at risk varient (402H) | | Descriptor: | COMPLEMENT FACTOR H | | Authors: | Herbert, A.P, Deakin, J.A, Schmidt, C.Q, Blaum, B.S, Egan, C, Ferreira, V.P, Pangburn, M.K, Lyon, M, Uhrin, D, Barlow, P.N. | | Deposit date: | 2007-02-16 | | Release date: | 2007-03-20 | | Last modified: | 2018-05-02 | | Method: | SOLUTION NMR | | Cite: | Structure shows that a glycosaminoglycan and protein recognition site in factor H is perturbed by age-related macular degeneration-linked single nucleotide polymorphism.
J. Biol. Chem., 282, 2007
|
|
3K1W
 
 | | New Classes of Potent and Bioavailable Human Renin Inhibitors | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetylamino-2-deoxy-alpha-L-idopyranose, 4-{4-[3-(2-bromo-5-fluorophenoxy)propyl]phenyl}-N-(2-chlorobenzyl)-N-cyclopropyl-1,2,5,6-tetrahydropyridine-3-carboxamide, ... | | Authors: | Prade, L. | | Deposit date: | 2009-09-29 | | Release date: | 2010-03-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | New classes of potent and bioavailable human renin inhibitors
Bioorg.Med.Chem.Lett., 19, 2009
|
|
2JFM
 
 | | CRYSTAL STRUCTURE OF HUMAN STE20-LIKE KINASE (UNLIGANDED FORM) | | Descriptor: | 1,2-ETHANEDIOL, STE20-LIKE SERINE-THREONINE KINASE | | Authors: | Pike, A.C.W, Rellos, P, Fedorov, O, Keates, T, Salah, E, Savitsky, P, Papagrigoriou, E, Bunkoczi, G, von Delft, F, Arrowsmith, C.H, Edwards, A, Weigelt, J, Sundstrom, M, Knapp, S. | | Deposit date: | 2007-02-02 | | Release date: | 2007-02-27 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Activation Segment Dimerization: A Mechanism for Kinase Autophosphorylation of Non-Consensus Sites.
Embo J., 27, 2008
|
|
2JWT
 
 | | Solution structure of Engrailed homeodomain WT | | Descriptor: | Segmentation polarity homeobox protein engrailed | | Authors: | Religa, T.L. | | Deposit date: | 2007-10-24 | | Release date: | 2008-04-01 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Comparison of multiple crystal structures with NMR data for engrailed homeodomain
J.Biomol.Nmr, 40, 2008
|
|
1A90
 
 | | RECOMBINANT MUTANT CHICKEN EGG WHITE CYSTATIN, NMR, 31 STRUCTURES | | Descriptor: | CYSTATIN | | Authors: | Dieckmann, T, Mitschang, L, Hofmann, M, Kos, J, Turk, V, Auerswald, E.A, Jaenicke, R, Oschkinat, H. | | Deposit date: | 1998-04-14 | | Release date: | 1998-06-17 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The structures of native phosphorylated chicken cystatin and of a recombinant unphosphorylated variant in solution.
J.Mol.Biol., 234, 1993
|
|
3K3I
 
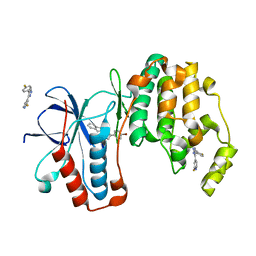 | | p38alpha bound to novel DGF-out compound PF-00215955 | | Descriptor: | (3S)-3-[4-(4-bromophenyl)-1H-imidazol-2-yl]-1,2,3,4-tetrahydroisoquinoline, 2-fluoro-4-[4-(4-fluorophenyl)-1H-pyrazol-3-yl]pyridine, Mitogen-activated protein kinase 14 | | Authors: | Kazmirski, S.L, DiNitto, J.P. | | Deposit date: | 2009-10-02 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The design, synthesis and potential utility of fluorescence probes that target DFG-out conformation of p38alpha for high throughput screening binding assay.
Chem.Biol.Drug Des., 74, 2009
|
|
1AI0
 
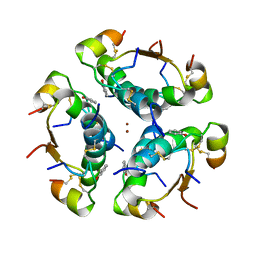 | | R6 HUMAN INSULIN HEXAMER (NON-SYMMETRIC), NMR, 10 STRUCTURES | | Descriptor: | PHENOL, R6 INSULIN HEXAMER, ZINC ION | | Authors: | Chang, X, Jorgensen, A.M.M, Bardrum, P, Led, J.J. | | Deposit date: | 1997-04-30 | | Release date: | 1997-11-12 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the R6 human insulin hexamer.
Biochemistry, 36, 1997
|
|
3K5R
 
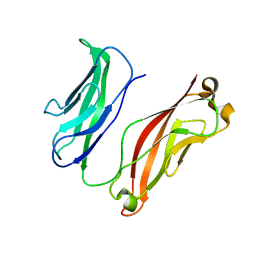 | |
2JS0
 
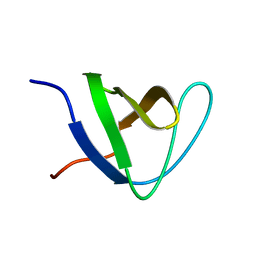 | |
1A08
 
 | |
3K6D
 
 | |
1A1J
 
 | | RADR (ZIF268 VARIANT) ZINC FINGER-DNA COMPLEX (GCGT SITE) | | Descriptor: | DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*CP*GP*T)-3'), DNA (5'-D(*TP*AP*CP*GP*CP*CP*CP*AP*CP*GP*C)-3'), PROTEIN (RADR ZIF268 ZINC FINGER PEPTIDE), ... | | Authors: | Elrod-Erickson, M, Benson, T.E, Pabo, C.O. | | Deposit date: | 1997-12-10 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution structures of variant Zif268-DNA complexes: implications for understanding zinc finger-DNA recognition.
Structure, 6, 1998
|
|
7F2F
 
 | | The complex of DNA with the C-terminal domain of TYE7 from Saccharomyces cerevisiae. | | Descriptor: | DNA (5'-D(*CP*AP*GP*AP*TP*CP*AP*TP*GP*TP*GP*TP*GP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*CP*AP*CP*AP*CP*AP*TP*GP*AP*TP*CP*T)-3'), Serine-rich protein TYE7 | | Authors: | Gui, W. | | Deposit date: | 2021-06-10 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of the complex of DNA with the C-terminal domain of TYE7 from Saccharomyces cerevisiae.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
2JXD
 
 | |
1A8G
 
 | | HIV-1 PROTEASE IN COMPLEX WITH SDZ283-910 | | Descriptor: | HIV-1 PROTEASE, benzyl [(1R)-1-({(1S,2S,3S)-1-benzyl-2-hydroxy-4-({(1S)-1-[(2-hydroxy-4-methoxybenzyl)carbamoyl]-2-methylpropyl}amino)-3-[(4-methoxybenzyl)amino]-4-oxobutyl}carbamoyl)-2,2-dimethylpropyl]carbamate | | Authors: | Kallen, J, Billich, A, Scholz, D, Auer, M, Kungl, A. | | Deposit date: | 1998-03-24 | | Release date: | 1998-07-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray structure and conformational dynamics of the HIV-1 protease in complex with the inhibitor SDZ283-910: agreement of time-resolved spectroscopy and molecular dynamics simulations.
J.Mol.Biol., 286, 1999
|
|
1A9U
 
 | | THE COMPLEX STRUCTURE OF THE MAP KINASE P38/SB203580 | | Descriptor: | 4-[5-(4-FLUORO-PHENYL)-2-(4-METHANESULFINYL-PHENYL)-3H-IMIDAZOL-4-YL]-PYRIDINE, MAP KINASE P38 | | Authors: | Wang, Z, Canagarajah, B, Boehm, J.C, Kassis, S, Cobb, M.H, Young, P.R, Abdel-Meguid, S, Adams, J.L, Goldsmith, E.J. | | Deposit date: | 1998-04-10 | | Release date: | 1999-04-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of inhibitor selectivity in MAP kinases.
Structure, 6, 1998
|
|
3JVR
 
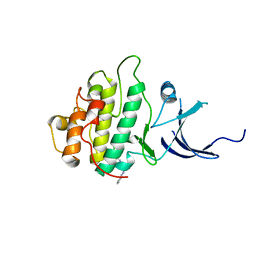 | | Characterization of the Chk1 allosteric inhibitor binding site | | Descriptor: | (1S)-1-(1H-benzimidazol-2-yl)ethyl (3,4-dichlorophenyl)carbamate, Serine/threonine-protein kinase Chk1 | | Authors: | Chen, P. | | Deposit date: | 2009-09-17 | | Release date: | 2009-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Characterization of the CHK1 allosteric inhibitor binding site.
Biochemistry, 48, 2009
|
|
2JDG
 
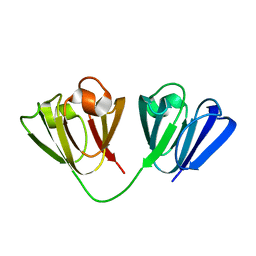 | | Affilin based on HUMAN GAMMA-B CRYSTALLIN | | Descriptor: | GAMMA CRYSTALLIN B | | Authors: | Ebersbach, H, Fiedler, E, Scheuermann, T, Fiedler, M, Stubbs, M.T, Reimann, C, Proetzel, G, Rudolph, R, Fiedler, U. | | Deposit date: | 2007-01-08 | | Release date: | 2007-07-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Affilin-Novel Binding Molecules Based on Human Gamma-B-Crystallin, an All Beta-Sheet Protein.
J.Mol.Biol., 372, 2007
|
|
1ABO
 
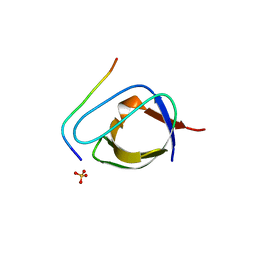 | | CRYSTAL STRUCTURE OF THE COMPLEX OF THE ABL TYROSINE KINASE SH3 DOMAIN WITH 3BP-1 SYNTHETIC PEPTIDE | | Descriptor: | 3BP-1 SYNTHETIC PEPTIDE, 10 RESIDUES, ABL TYROSINE KINASE, ... | | Authors: | Musacchio, A, Wilmanns, M, Saraste, M. | | Deposit date: | 1995-05-19 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution crystal structures of tyrosine kinase SH3 domains complexed with proline-rich peptides.
Nat.Struct.Biol., 1, 1994
|
|
2JK1
 
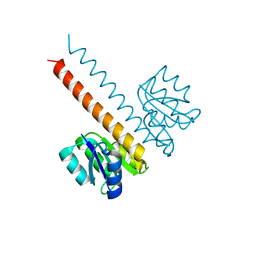 | | Crystal structure of the wild-type HupR receiver domain | | Descriptor: | HYDROGENASE TRANSCRIPTIONAL REGULATORY PROTEIN HUPR1, MAGNESIUM ION | | Authors: | Davies, K.M, Lowe, E.D, Venien-Bryan, C, Johnson, L.N. | | Deposit date: | 2008-05-26 | | Release date: | 2008-11-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Hupr Receiver Domain Crystal Structure in its Nonphospho and Inhibitory Phospho States.
J.Mol.Biol., 385, 2009
|
|
3JUS
 
 | | Crystal structure of human lanosterol 14alpha-demethylase (CYP51) in complex with econazole | | Descriptor: | 1-[(2R)-2-[(4-chlorobenzyl)oxy]-2-(2,4-dichlorophenyl)ethyl]-1H-imidazole, 1-[(2S)-2-[(4-CHLOROBENZYL)OXY]-2-(2,4-DICHLOROPHENYL)ETHYL]-1H-IMIDAZOLE, Cycloheptakis-(1-4)-(alpha-D-glucopyranose), ... | | Authors: | Strushkevich, N, MacKenzie, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Usanov, S.A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-15 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of human CYP51 inhibition by antifungal azoles.
J. Mol. Biol., 397, 2010
|
|
3JSD
 
 | | Insulin's biosynthesis and activity have opposing structural requirements: a new factor in neonatal diabetes mellitus | | Descriptor: | CHLORIDE ION, Insulin A chain, Insulin B chain, ... | | Authors: | Weiss, M.A, Wan, Z.L, Dodson, E.J, Liu, M, Xu, B, Hua, Q.X, Turkenburg, M, Whittingham, J, Nakagawa, S.H, Huang, K, Hu, S.Q, Jia, W.H, Wang, S.H, Brange, J, Whittaker, J, Arvan, P, Katsoyannis, P.G, Dodson, G.G. | | Deposit date: | 2009-09-10 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insulin's biosynthesis and activity have opposing structural requirements: a new factor in neonatal diabetes mellitus
To be Published
|
|
1AN2
 
 | | RECOGNITION BY MAX OF ITS COGNATE DNA THROUGH A DIMERIC B/HLH/Z DOMAIN | | Descriptor: | DNA (5'-D(*GP*TP*GP*TP*AP*GP*GP*TP*CP*AP*CP*GP*TP*GP*AP*CP*C P*TP*AP*CP*AP*C)- 3'), PROTEIN (TRANSCRIPTION FACTOR MAX (TF MAX)) | | Authors: | Ferre-D'Amare, A.R, Prendergast, G.C, Ziff, E.B, Burley, S.K. | | Deposit date: | 1996-09-06 | | Release date: | 1997-09-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Recognition by Max of its cognate DNA through a dimeric b/HLH/Z domain.
Nature, 363, 1993
|
|
3K3J
 
 | | P38alpha bound to novel DFG-out compound PF-00416121 | | Descriptor: | 2-(4-fluorophenyl)-3-oxo-6-pyridin-4-yl-N-[2-(trifluoromethyl)benzyl]-2,3-dihydropyridazine-4-carboxamide, 2-fluoro-4-[4-(4-fluorophenyl)-1H-pyrazol-3-yl]pyridine, Mitogen-activated protein kinase 14 | | Authors: | Kazmirski, S.L, DiNitto, J.P. | | Deposit date: | 2009-10-02 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | The Design, Synthesis and Potential Utility of Fluorescence Probes that Target DFG-out Conformation of p38alpha for High Throughput Screening Binding Assay.
Chem.Biol.Drug Des., 74, 2009
|
|
1AFK
 
 | |
