456D
 
 | | MOLECULAR AND CRYSTAL STRUCTURE OF D(CGCGMO6AATCCGCG): THE WATSON-CRICK TYPE N6-METHOXYADENOSINE/CYTIDINE BASE-PAIRS IN B-DNA | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*(A47)P*AP*TP*CP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Chatake, T, Ono, A, Ueno, Y, Matsuda, A, Takenaka, A. | | Deposit date: | 1999-03-06 | | Release date: | 2000-01-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic studies on damaged DNAs. I. An N(6)-methoxyadenine residue forms a Watson-Crick pair with a cytosine residue in a B-DNA duplex.
J.Mol.Biol., 294, 1999
|
|
457D
 
 | | MOLECULAR AND CRYSTAL STRUCTURE OF D(CGCGMO6AATTCGCG): N6-METHOXYADENOSINE/ THYMIDINE BASE-PAIRS IN B-DNA | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*(A47)P*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Chatake, T, Ono, A, Ueno, Y, Matsuda, A, Takenaka, A. | | Deposit date: | 1999-03-06 | | Release date: | 2000-01-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic studies on damaged DNAs. II. N(6)-methoxyadenine can present two alternate faces for Watson-Crick base-pairing, leading to pyrimidine transition mutagenesis.
J.Mol.Biol., 294, 1999
|
|
1QQT
 
 | | METHIONYL-TRNA SYNTHETASE FROM ESCHERICHIA COLI | | Descriptor: | METHIONYL-TRNA SYNTHETASE, ZINC ION | | Authors: | Mechulam, Y, Schmitt, E, Maveyraud, L, Zelwer, C, Nureki, O, Yokoyama, S, Konno, M, Blanquet, S. | | Deposit date: | 1999-06-08 | | Release date: | 2000-01-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure of Escherichia coli methionyl-tRNA synthetase highlights species-specific features.
J.Mol.Biol., 294, 1999
|
|
1CJG
 
 | | NMR STRUCTURE OF LAC REPRESSOR HP62-DNA COMPLEX | | Descriptor: | DNA (5'-D(*GP*AP*AP*TP*TP*GP*TP*GP*AP*GP*CP*GP*CP*TP*CP*AP*CP*AP*AP*TP*TP*C)-3'), PROTEIN (LAC REPRESSOR) | | Authors: | Spronk, C.A.E.M, Bonvin, A.M.J.J, Radha, P.K, Melacini, G, Boelens, R, Kaptein, R. | | Deposit date: | 1999-04-14 | | Release date: | 2000-01-01 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The solution structure of Lac repressor headpiece 62 complexed to a symmetrical lac operator.
Structure Fold.Des., 7, 1999
|
|
1CL3
 
 | | MOLECULAR INSIGHTS INTO PEBP2/CBF-SMMHC ASSOCIATED ACUTE LEUKEMIA REVEALED FROM THE THREE-DIMENSIONAL STRUCTURE OF PEBP2/CBF BETA | | Descriptor: | POLYOMAVIRUS ENHANCER BINDING PROTEIN 2 | | Authors: | Goger, M, Gupta, V, Kim, W.Y, Shigesada, K, Ito, Y, Werner, M.H. | | Deposit date: | 1999-05-04 | | Release date: | 2000-01-01 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Molecular insights into PEBP2/CBF beta-SMMHC associated acute leukemia revealed from the structure of PEBP2/CBF beta
Nat.Struct.Biol., 6, 1999
|
|
1QFN
 
 | |
1D3R
 
 | | CRYSTAL STRUCTURE OF TRIPLEX DNA | | Descriptor: | DNA (5'-D(*CP*(BRU)P*CP*CP*(BRU)P*CP*CP*GP*CP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*GP*CP*GP*GP*AP*G)-3') | | Authors: | Rhee, S, Han, Z.-J, Liu, K, Todd Miles, H.T, Davies, D.R. | | Deposit date: | 1999-09-30 | | Release date: | 2000-01-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a triple helical DNA with a triplex-duplex junction.
Biochemistry, 38, 1999
|
|
1QR1
 
 | | POOR BINDING OF A HER-2/NEU EPITOPE (GP2) TO HLA-A2.1 IS DUE TO A LACK OF INTERACTIONS IN THE CENTER OF THE PEPTIDE | | Descriptor: | BETA-2 MICROGLOBULIN, GP2 PEPTIDE, HLA-A2.1 HEAVY CHAIN | | Authors: | Kuhns, J.J, Batalia, M.A, Yan, S, Collins, E.J. | | Deposit date: | 1999-06-17 | | Release date: | 2000-01-01 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Poor binding of a HER-2/neu epitope (GP2) to HLA-A2.1 is due to a lack of interactions with the center of the peptide.
J.Biol.Chem., 274, 1999
|
|
1QLH
 
 | |
1QLJ
 
 | |
1DX6
 
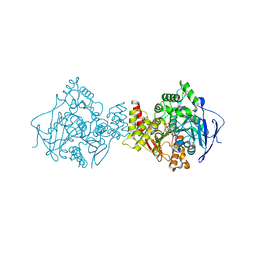 | | STRUCTURE OF ACETYLCHOLINESTERASE COMPLEXED WITH (-)-GALANTHAMINE AT 2.3A RESOLUTION | | Descriptor: | (-)-GALANTHAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Greenblatt, H.M, Kryger, G, Lewis, T.T, Silman, I, Sussman, J.L. | | Deposit date: | 1999-12-21 | | Release date: | 2000-01-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Acetylcholinesterase Complexed with (-)-Galanthamine at 2.3A Resolution
FEBS Lett., 463, 1999
|
|
1DPW
 
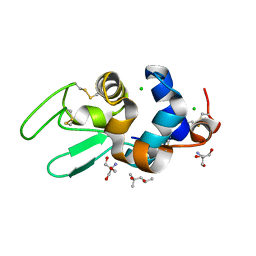 | | STRUCTURE OF HEN EGG-WHITE LYSOZYME IN COMPLEX WITH MPD | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Weiss, M.S, Palm, G.J, Hilgenfeld, R. | | Deposit date: | 1999-12-28 | | Release date: | 2000-01-03 | | Last modified: | 2011-11-23 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystallization, structure solution and refinement of hen egg-white lysozyme at pH 8.0 in the presence of MPD.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1DPX
 
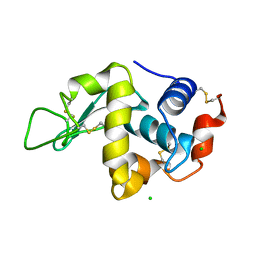 | | STRUCTURE OF HEN EGG-WHITE LYSOZYME | | Descriptor: | CHLORIDE ION, LYSOZYME | | Authors: | Weiss, M.S, Palm, G.J, Hilgenfeld, R. | | Deposit date: | 1999-12-28 | | Release date: | 2000-01-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystallization, structure solution and refinement of hen egg-white lysozyme at pH 8.0 in the presence of MPD.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1CZ1
 
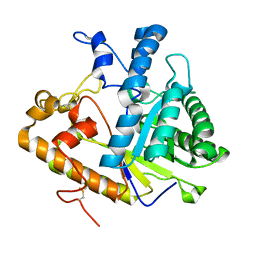 | | EXO-B-(1,3)-GLUCANASE FROM CANDIDA ALBICANS AT 1.85 A RESOLUTION | | Descriptor: | PROTEIN (EXO-B-(1,3)-GLUCANASE) | | Authors: | Cutfield, S.M, Davies, G.J, Murshudov, G, Anderson, B.F, Moody, P.C.E, Sullivan, P.A, Cutfield, J.F. | | Deposit date: | 1999-09-01 | | Release date: | 2000-01-03 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The structure of the exo-beta-(1,3)-glucanase from Candida albicans in native and bound forms: relationship between a pocket and groove in family 5 glycosyl hydrolases.
J.Mol.Biol., 294, 1999
|
|
1DCF
 
 | |
1DX8
 
 | | Rubredoxin from Guillardia theta | | Descriptor: | RUBREDOXIN, ZINC ION | | Authors: | Schweimer, K, Hoffmann, S, Wastl, J, Maier, U.G, Roesch, P, Sticht, H. | | Deposit date: | 1999-12-23 | | Release date: | 2000-01-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a zinc substituted eukaryotic rubredoxin from the cryptomonad alga Guillardia theta.
Protein Sci., 9, 2000
|
|
1QGX
 
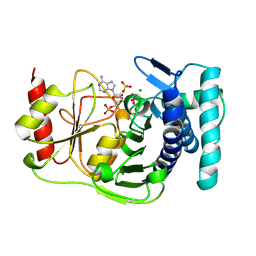 | | X-RAY STRUCTURE OF YEAST HAL2P | | Descriptor: | 3',5'-ADENOSINE BISPHOSPHATASE, ADENOSINE MONOPHOSPHATE, BETA-MERCAPTOETHANOL, ... | | Authors: | Albert, A, Yenush, L, Gil-Mascarell, M.R, Rodriguez, P.L, Patel, J, Martinez-Ripoll, M, Blundell, T.L, Serrano, R. | | Deposit date: | 1999-05-10 | | Release date: | 2000-01-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray structure of yeast Hal2p, a major target of lithium and sodium toxicity, and identification of framework interactions determining cation sensitivity.
J.Mol.Biol., 295, 2000
|
|
1DC7
 
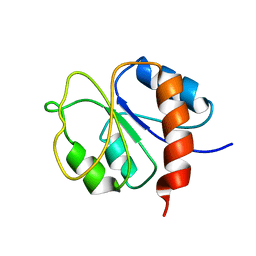 | | STRUCTURE OF A TRANSIENTLY PHOSPHORYLATED "SWITCH" IN BACTERIAL SIGNAL TRANSDUCTION | | Descriptor: | NITROGEN REGULATION PROTEIN | | Authors: | Kern, D, Volkman, B.F, Luginbuhl, P, Nohaile, M.J, Kustu, S, Wemmer, D.E. | | Deposit date: | 1999-11-04 | | Release date: | 2000-01-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of a transiently phosphorylated switch in bacterial signal transduction.
Nature, 402, 1999
|
|
1DC8
 
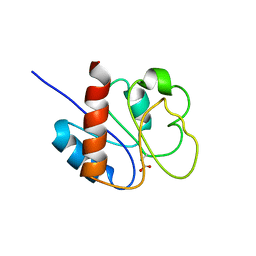 | | STRUCTURE OF A TRANSIENTLY PHOSPHORYLATED "SWITCH" IN BACTERIAL SIGNAL TRANSDUCTION | | Descriptor: | NITROGEN REGULATION PROTEIN | | Authors: | Kern, D, Volkman, B.F, Luginbuhl, P, Nohaile, M.J, Kustu, S, Wemmer, D.E. | | Deposit date: | 1999-11-04 | | Release date: | 2000-01-05 | | Last modified: | 2022-12-21 | | Method: | SOLUTION NMR | | Cite: | Structure of a transiently phosphorylated switch in bacterial signal transduction.
Nature, 402, 1999
|
|
1DNL
 
 | | X-RAY STRUCTURE OF ESCHERICHIA COLI PYRIDOXINE 5'-PHOSPHATE OXIDASE COMPLEXED WITH FMN AT 1.8 ANGSTROM RESOLUTION | | Descriptor: | FLAVIN MONONUCLEOTIDE, PHOSPHATE ION, PYRIDOXINE 5'-PHOSPHATE OXIDASE | | Authors: | Safo, M.K, Mathews, I, Musayev, F.N, di Salvo, M.L, Thiel, D.J, Abraham, D.J, Schirch, V. | | Deposit date: | 1999-12-16 | | Release date: | 2000-01-05 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray structure of Escherichia coli pyridoxine 5'-phosphate oxidase complexed with FMN at 1.8 A resolution.
Structure Fold.Des., 8, 2000
|
|
1B4O
 
 | |
6ALD
 
 | | RABBIT MUSCLE ALDOLASE A/FRUCTOSE-1,6-BISPHOSPHATE COMPLEX | | Descriptor: | 1,6-FRUCTOSE DIPHOSPHATE (LINEAR FORM), FRUCTOSE-1,6-BIS(PHOSPHATE) ALDOLASE | | Authors: | Choi, K.H, Mazurkie, A.S, Morris, A.J, Utheza, D, Tolan, D.R, Allen, K.N. | | Deposit date: | 1998-12-23 | | Release date: | 2000-01-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a fructose-1,6-bis(phosphate) aldolase liganded to its natural substrate in a cleavage-defective mutant at 2.3 A(,).
Biochemistry, 38, 1999
|
|
1BK8
 
 | |
1C2T
 
 | | NEW INSIGHTS INTO INHIBITOR DESIGN FROM THE CRYSTAL STRUCTURE AND NMR STUDIES OF E. COLI GAR TRANSFORMYLASE IN COMPLEX WITH BETA-GAR AND 10-FORMYL-5,8,10-TRIDEAZAFOLIC ACID. | | Descriptor: | 10-FORMYL-5,8,10-TRIDEAZAFOLIC ACID, GLYCINAMIDE RIBONUCLEOTIDE, GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE | | Authors: | Greasley, S.E, Yamashita, M.M, Cai, H, Benkovic, S.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 1999-07-26 | | Release date: | 2000-01-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New insights into inhibitor design from the crystal structure and NMR studies of Escherichia coli GAR transformylase in complex with beta-GAR and 10-formyl-5,8,10-trideazafolic acid.
Biochemistry, 38, 1999
|
|
1DOQ
 
 | |
