3EIQ
 
 | | Crystal structure of Pdcd4-eIF4A | | Descriptor: | Eukaryotic initiation factor 4A-I, Programmed cell death protein 4 | | Authors: | Loh, P.G, Cheng, Z, Song, H. | | Deposit date: | 2008-09-17 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for translational inhibition by the tumour suppressor Pdcd4
Embo J., 28, 2009
|
|
5L3D
 
 | | Human LSD1/CoREST: LSD1 Y761H mutation | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1A, REST corepressor 1 | | Authors: | Pilotto, S, Speranzini, V, Marabelli, C, Mattevi, A. | | Deposit date: | 2016-04-06 | | Release date: | 2016-05-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | LSD1/KDM1A mutations associated to a newly described form of intellectual disability impair demethylase activity and binding to transcription factors.
Hum.Mol.Genet., 25, 2016
|
|
3V2U
 
 | | Crystal structure of the yeast GAL regulon complex of the repressor, Gal80p, and the transducer, Gal3p, with galactose and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, Galactose/lactose metabolism regulatory protein GAL80, ... | | Authors: | Lavy, T, Kumar, P.R, He, H, Joshua-Tor, L. | | Deposit date: | 2011-12-12 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | The Gal3p transducer of the GAL regulon interacts with the Gal80p repressor in its ligand-induced closed conformation.
Genes Dev., 26, 2012
|
|
5L3B
 
 | | Human LSD1/CoREST: LSD1 D556G mutation | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1A, REST corepressor 1 | | Authors: | Pilotto, S, Speranzini, V, Marabelli, C. | | Deposit date: | 2016-04-06 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | LSD1/KDM1A mutations associated to a newly described form of intellectual disability impair demethylase activity and binding to transcription factors.
Hum.Mol.Genet., 25, 2016
|
|
5BUJ
 
 | | ERK2 complexed with a N-H tetrahydroazaindazole | | Descriptor: | 4-[3-(pyridin-4-yl)-2,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl]pyridin-2(1H)-one, Mitogen-activated protein kinase 1 | | Authors: | Bellamacina, C.R, Shu, W, Bussiere, D.E, Bagdanoff, J.T. | | Deposit date: | 2015-06-03 | | Release date: | 2015-07-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Ligand efficient tetrahydro-pyrazolopyridines as inhibitors of ERK2 kinase.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
4H9S
 
 | | Complex structure 6 of DAXX/H3.3(sub7)/H4 | | Descriptor: | Death domain-associated protein 6, Histone H3.3, Histone H4, ... | | Authors: | Elsasser, S.J, Huang, H, Lewis, P.W, Chin, J.W, Allis, D.C, Patel, D.J. | | Deposit date: | 2012-09-24 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | DAXX chaperone envelops an H3.3/H4 dimer dictating H3.3-specific read out
To be Published
|
|
4BKX
 
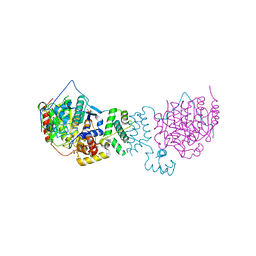 | | The structure of HDAC1 in complex with the dimeric ELM2-SANT domain of MTA1 from the NuRD complex | | Descriptor: | ACETATE ION, HISTONE DEACETYLASE 1, METASTASIS-ASSOCIATED PROTEIN MTA1, ... | | Authors: | Millard, C.J, Watson, P.J, Celardo, I, Gordiyenko, Y, Cowley, S.M, Robinson, C.V, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2013-04-30 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Class I Hdacs Share a Common Mechanism of Regulation by Inositol Phosphates.
Mol.Cell, 51, 2013
|
|
2A7O
 
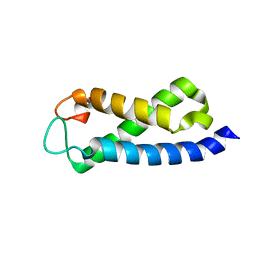 | | Solution Structure of the hSet2/HYPB SRI domain | | Descriptor: | Huntingtin interacting protein B | | Authors: | Li, M, Phatnani, H.P, Guan, Z, Sage, H, Greenleaf, A, Zhou, P. | | Deposit date: | 2005-07-05 | | Release date: | 2005-11-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Set2 Rpb1 interacting domain of human Set2 and its interaction with the hyperphosphorylated C-terminal domain of Rpb1
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
3AV2
 
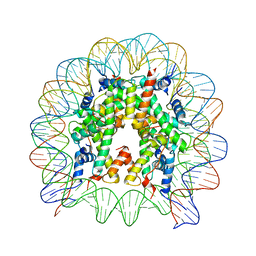 | | The human nucleosome structure containing the histone variant H3.3 | | Descriptor: | 146-MER DNA, Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Tachiwana, H, Osakabe, A, Shiga, T, Miya, M, Kimura, H, Kagawa, W, Kurumizaka, H. | | Deposit date: | 2011-02-18 | | Release date: | 2011-06-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of human nucleosomes containing major histone H3 variants
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3ZMS
 
 | | LSD1-CoREST in complex with INSM1 peptide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, INSULINOMA-ASSOCIATED PROTEIN 1, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, ... | | Authors: | Tortorici, M, Borrello, M.T, Tardugno, M, Chiarelli, L.R, Pilotto, S, Ciossani, G, Vellore, N.A, Cowan, J, O'Connell, M, Mai, A, Baron, R, Ganesan, A, Mattevi, A. | | Deposit date: | 2013-02-12 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Protein Recognition by Small Peptide Reversible Inhibitors of the Chromatin-Modifying Lsd1/Corest Lysine Demethylase.
Acs Chem.Biol., 8, 2013
|
|
4J2X
 
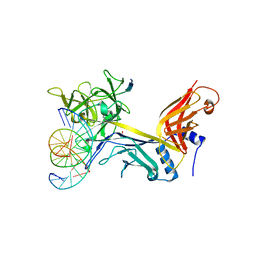 | | CSL (RBP-Jk) with corepressor KyoT2 bound to DNA | | Descriptor: | 1,2-ETHANEDIOL, 5'-D(*AP*AP*TP*CP*TP*TP*TP*CP*CP*CP*AP*CP*AP*GP*T)-3', 5'-D(*TP*TP*AP*CP*TP*GP*TP*GP*GP*GP*AP*AP*AP*GP*A)-3', ... | | Authors: | Collins, K.J, Kovall, R.A. | | Deposit date: | 2013-02-05 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure and Function of the CSL-KyoT2 Corepressor Complex: A Negative Regulator of Notch Signaling.
Structure, 22, 2014
|
|
4B6E
 
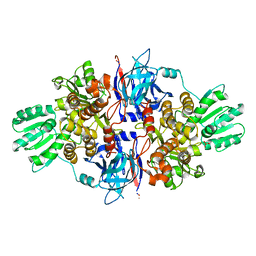 | | Discovery of an allosteric mechanism for the regulation of HCV NS3 protein function | | Descriptor: | 1H-indazol-7-amine, NON-STRUCTURAL PROTEIN 4A, SERINE PROTEASE NS3, ... | | Authors: | Saalau-Bethell, S.M, Woodhead, A.J, Chessari, G, Carr, M.G, Coyle, J, Graham, B, Hiscock, S.D, Murray, C.W, Pathuri, P, Rich, S.J, Richardson, C.J, Williams, P.A, Jhoti, H. | | Deposit date: | 2012-08-09 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Discovery of an Allosteric Mechanism for the Regulation of Hcv Ns3 Protein Function.
Nat.Chem.Biol., 8, 2012
|
|
1LWE
 
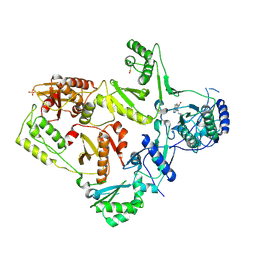 | | CRYSTAL STRUCTURE OF M41L/T215Y MUTANT HIV-1 REVERSE TRANSCRIPTASE (RTMN) IN COMPLEX WITH NEVIRAPINE | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 REVERSE TRANSCRIPTASE, PHOSPHATE ION | | Authors: | Ren, J, Chamberlain, P.P, Nichols, C.E, Douglas, L, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2002-05-31 | | Release date: | 2002-10-30 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Crystal structures of Zidovudine- or Lamivudine-resistant human immunodeficiency virus type 1 reverse transcriptases containing mutations at codons 41, 184, and 215.
J.Virol., 76, 2002
|
|
4B75
 
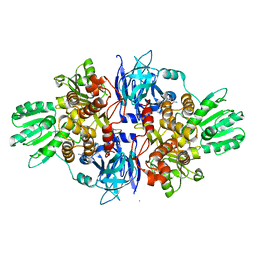 | | Discovery of an allosteric mechanism for the regulation of HCV NS3 protein function | | Descriptor: | (2S)-4-amino-N-[(1R)-1-(4-chloro-2-fluoro-3-phenoxyphenyl)propyl]-4-oxobutan-2-aminium, NON-STRUCTURAL PROTEIN 4A, SERINE PROTEASE NS3 | | Authors: | Saalau-Bethell, S.M, Woodhead, A.J, Chessari, G, Carr, M.G, Coyle, J, Graham, B, Hiscock, S.D, Murray, C.W, Pathuri, P, Rich, S.J, Richardson, C.J, Williams, P.A, Jhoti, H. | | Deposit date: | 2012-08-16 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Discovery of an Allosteric Mechanism for the Regulation of Hcv Ns3 Protein Function.
Nat.Chem.Biol., 8, 2012
|
|
8CDK
 
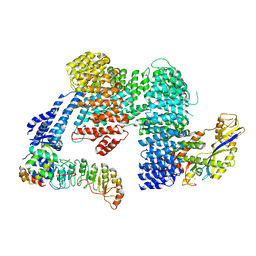 | |
8CDJ
 
 | | CAND1 b-hairpin++-SCF-SKP2 CAND1 rolling SCF engaged | | Descriptor: | Cullin-1, Cullin-associated NEDD8-dissociated protein 1, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Baek, K, Schulman, B.A. | | Deposit date: | 2023-01-31 | | Release date: | 2023-04-19 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Systemwide disassembly and assembly of SCF ubiquitin ligase complexes.
Cell, 186, 2023
|
|
5FWM
 
 | | Atomic cryoEM structure of Hsp90-Cdc37-Cdk4 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CYCLIN-DEPENDENT KINASE 4, HEAT SHOCK PROTEIN HSP 90 BETA, ... | | Authors: | Verba, K.A, Wang, R.Y.R, Arakawa, A, Liu, Y, Yokoyama, S, Agard, D.A. | | Deposit date: | 2016-02-18 | | Release date: | 2016-07-06 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Atomic Structure of Hsp90-Cdc37-Cdk4 Reveals that Hsp90 Traps and Stabilizes an Unfolded Kinase.
Science, 352, 2016
|
|
4B74
 
 | | Discovery of an allosteric mechanism for the regulation of HCV NS3 protein function | | Descriptor: | (2S)-4-[(2-ammonioethyl)amino]-N-[(1R)-1-(4-chloro-2-fluoro-3-phenoxyphenyl)propyl]-4-oxobutan-2-aminium, NON-STRUCTURAL PROTEIN 4A, SERINE PROTEASE NS3 | | Authors: | Saalau-Bethell, S.M, Woodhead, A.J, Chessari, G, Carr, M.G, Coyle, J, Graham, B, Hiscock, S.D, Murray, C.W, Pathuri, P, Rich, S.J, Richardson, C.J, Williams, P.A, Jhoti, H. | | Deposit date: | 2012-08-16 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Discovery of an Allosteric Mechanism for the Regulation of Hcv Ns3 Protein Function
Nat.Chem.Biol., 8, 2012
|
|
8CQL
 
 | | pVHL:EloB:EloC in complex with (2S,4R)-N-((S)-1-(5-Fluoro-2-methoxy-4-(4-methylthiazol-5-yl)phenyl)ethyl)-1-((S)-2-(1-fluorocyclopropane-1-carboxamido)-3,3-dimethylbutanoyl)-4-hydroxypyrrolidine-2-carboxamide (Compound 33) | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-2-[(1-fluoranylcyclopropyl)carbonylamino]-3,3-dimethyl-butanoyl]-~{N}-[(1~{S})-1-[5-fluoranyl-2-methoxy-4-(4-methyl-1,3-thiazol-5-yl)phenyl]ethyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Casement, R, Phuong Vu, L, Ciulli, A, Gutschow, M. | | Deposit date: | 2023-03-06 | | Release date: | 2023-09-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Expanding the Structural Diversity at the Phenylene Core of Ligands for the von Hippel-Lindau E3 Ubiquitin Ligase: Development of Highly Potent Hypoxia-Inducible Factor-1 alpha Stabilizers.
J.Med.Chem., 66, 2023
|
|
8CQK
 
 | | pVHL:EloB:EloC in complex with (2S,4R)-1-((S)-2-(1-Fluorocyclopropane-1-carboxamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-((S)-1-(2-methyl-4-(4-methylthiazol-5-yl)phenyl)ethyl)pyrrolidine-2-carboxamide (Compound 30) | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-2-[(1-fluoranylcyclopropyl)carbonylamino]-3,3-dimethyl-butanoyl]-~{N}-[(1~{S})-1-[2-methyl-4-(4-methyl-1,3-thiazol-5-yl)phenyl]ethyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Casement, R, Phuong Vu, L, Ciulli, A, Gutschow, M. | | Deposit date: | 2023-03-06 | | Release date: | 2023-09-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Expanding the Structural Diversity at the Phenylene Core of Ligands for the von Hippel-Lindau E3 Ubiquitin Ligase: Development of Highly Potent Hypoxia-Inducible Factor-1 alpha Stabilizers.
J.Med.Chem., 66, 2023
|
|
8CQE
 
 | | pVHL:EloB:EloC in complex with (2S,4R)-1-((S)-2-(1-Fluorocyclopropane-1-carboxamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-((S)-1-(2-methyl-4-(4-methylthiazol-5-yl)phenyl)ethyl)pyrrolidine-2-carboxamide (Compound 37) | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-2-[(1-fluoranylcyclopropyl)carbonylamino]-3,3-dimethyl-butanoyl]-~{N}-[(1~{S})-7-fluoranyl-6-(4-methyl-1,3-thiazol-5-yl)-1,2,3,4-tetrahydronaphthalen-1-yl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Casement, R, Phuong Vu, L, Ciulli, A, Gutschow, M. | | Deposit date: | 2023-03-06 | | Release date: | 2023-09-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Expanding the Structural Diversity at the Phenylene Core of Ligands for the von Hippel-Lindau E3 Ubiquitin Ligase: Development of Highly Potent Hypoxia-Inducible Factor-1 alpha Stabilizers.
J.Med.Chem., 66, 2023
|
|
4B76
 
 | | Discovery of an allosteric mechanism for the regulation of HCV NS3 protein function | | Descriptor: | NON-STRUCTURAL PROTEIN 4A, SERINE PROTEASE NS3, [2,4-bis(fluoranyl)-3-phenoxy-phenyl]methylazanium | | Authors: | Saalau-Bethell, S.M, Woodhead, A.J, Chessari, G, Carr, M.G, Coyle, J, Graham, B, Hiscock, S.D, Murray, C.W, Pathuri, P, Rich, S.J, Richardson, C.J, Williams, P.A, Jhoti, H. | | Deposit date: | 2012-08-16 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Discovery of an Allosteric Mechanism for the Regulation of Hcv Ns3 Protein Function.
Nat.Chem.Biol., 8, 2012
|
|
6BOR
 
 | | Human APE1 substrate complex with an G/G mismatch adjacent the THF | | Descriptor: | 1,2-ETHANEDIOL, 21-mer DNA, CHLORIDE ION, ... | | Authors: | Freudenthal, B.D, Whitaker, A.M, Fairlamb, M.S. | | Deposit date: | 2017-11-20 | | Release date: | 2018-08-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Apurinic/apyrimidinic (AP) endonuclease 1 processing of AP sites with 5' mismatches.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6BOT
 
 | | Human APE1 substrate complex with an C/C mismatch adjacent the THF | | Descriptor: | 1,2-ETHANEDIOL, 21-mer DNA, DNA-(apurinic or apyrimidinic site) lyase | | Authors: | Freudenthal, B.D, Whitaker, A.M, Fairlamb, M.S. | | Deposit date: | 2017-11-20 | | Release date: | 2018-08-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Apurinic/apyrimidinic (AP) endonuclease 1 processing of AP sites with 5' mismatches.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
1LW2
 
 | | CRYSTAL STRUCTURE OF T215Y MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH 1051U91 | | Descriptor: | 6,11-DIHYDRO-11-ETHYL-6-METHYL-9-NITRO-5H-PYRIDO[2,3-B][1,5]BENZODIAZEPIN-5-ONE, HIV-1 REVERSE TRANSCRIPTASE, PHOSPHATE ION | | Authors: | Ren, J, Chamberlain, P.P, Nichols, C.E, Douglas, L, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2002-05-30 | | Release date: | 2002-10-30 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of Zidovudine- or Lamivudine-resistant human immunodeficiency virus type 1 reverse transcriptases containing mutations at codons 41, 184, and 215.
J.Virol., 76, 2002
|
|
