6FBB
 
 | | Crystal structure of 14-3-3 sigma in complex with wild-type Shroom3 | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Leysen, S, Meijer, F.A, Milroy, L.G, Ottmann, C. | | Deposit date: | 2017-12-18 | | Release date: | 2018-03-14 | | Last modified: | 2018-05-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Characterization of Coding/Noncoding Variants forSHROOM3in Patients with CKD.
J. Am. Soc. Nephrol., 29, 2018
|
|
6FAW
 
 | | Crystal structure of C-terminal modified Tau peptide-hybrid 4.2c-I with 14-3-3sigma | | Descriptor: | (2~{R})-2-[(~{R})-[2-(2-methoxyethoxy)phenyl]-phenyl-methyl]pyrrolidine, 14-3-3 protein sigma, ACE-ARG-THR-PRO-SEP-LEU-PRO-GLY, ... | | Authors: | Andrei, S.A, Meijer, F.A, Ottmann, C, Milroy, L.G. | | Deposit date: | 2017-12-18 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Inhibition of 14-3-3/Tau by Hybrid Small-Molecule Peptides Operating via Two Different Binding Modes.
ACS Chem Neurosci, 9, 2018
|
|
3J82
 
 | | Electron cryo-microscopy of DNGR-1 in complex with F-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Hanc, P, Fujii, T, Yamada, Y, Huotari, J, Schulz, O, Ahrens, S, Kjaer, S, Way, M, Namba, K, Reis e Sousa, C. | | Deposit date: | 2014-09-25 | | Release date: | 2015-05-20 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Structure of the Complex of F-Actin and DNGR-1, a C-Type Lectin Receptor Involved in Dendritic Cell Cross-Presentation of Dead Cell-Associated Antigens.
Immunity, 42, 2015
|
|
3ZN0
 
 | | LSD1-CoREST in complex with PRSFAA peptide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, PEPTIDE, ... | | Authors: | Tortorici, M, Borrello, M.T, Tardugno, M, Chiarelli, L.R, Pilotto, S, Ciossani, G, Vellore, N.A, Cowan, J, O'Connell, M, Mai, A, Baron, R, Ganesan, A, Mattevi, A. | | Deposit date: | 2013-02-13 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Protein Recognition by Small Peptide Reversible Inhibitors of the Chromatin-Modifying Lsd1/Corest Lysine Demethylase.
Acs Chem.Biol., 8, 2013
|
|
3ZMZ
 
 | | LSD1-CoREST in complex with PRSFAV peptide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, PEPTIDE, ... | | Authors: | Tortorici, M, Borrello, M.T, Tardugno, M, Chiarelli, L.R, Pilotto, S, Ciossani, G, Vellore, N.A, Cowan, J, O'Connell, M, Mai, A, Baron, R, Ganesan, A, Mattevi, A. | | Deposit date: | 2013-02-13 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Protein Recognition by Small Peptide Reversible Inhibitors of the Chromatin-Modifying Lsd1/Corest Lysine Demethylase.
Acs Chem.Biol., 8, 2013
|
|
3L1Z
 
 | |
2ZR6
 
 | |
2Z7F
 
 | |
3KAF
 
 | | Structure-guided design of alpha-amino acid-derived Pin1 inhibitors | | Descriptor: | 3-(1H-benzimidazol-2-yl)-N-(1-benzothiophen-2-ylcarbonyl)-D-alanine, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Baker, L.M, Dokurno, P, Robinson, D.A, Surgenor, A.E, Murray, J.B, Potter, A.J, Moore, J.D. | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-guided design of alpha-amino acid-derived Pin1 inhibitors
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3UKU
 
 | | Structure of Arp2/3 complex with bound inhibitor CK-869 | | Descriptor: | (2S)-2-(3-bromophenyl)-3-(2,4-dimethoxyphenyl)-1,3-thiazolidin-4-one, ACTIN-LIKE PROTEIN 2, ACTIN-LIKE PROTEIN 3, ... | | Authors: | Nolen, B.J, Han, M. | | Deposit date: | 2011-11-09 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of Arp2/3 complex with bound inhibitor CK-869
To be Published
|
|
6FCP
 
 | | Crystal structure of 14-3-3 sigma in complex with Shroom3 P1244L | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Leysen, S, Meijer, F.A, Milroy, L.G, Ottmann, C. | | Deposit date: | 2017-12-20 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Characterization of Coding/Noncoding Variants forSHROOM3in Patients with CKD.
J. Am. Soc. Nephrol., 29, 2018
|
|
6FCV
 
 | | Structure of the human DDB1-CSA complex | | Descriptor: | DNA damage-binding protein 1, DNA excision repair protein ERCC-8 | | Authors: | Meulenbroek, E.M, Pannu, N.S. | | Deposit date: | 2017-12-21 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | TRiC controls transcription resumption after UV damage by regulating Cockayne syndrome protein A.
Nat Commun, 9, 2018
|
|
1JPY
 
 | | Crystal structure of IL-17F | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hymowitz, S.G, Filvaroff, E.H, Yin, J, Lee, J, Cai, L, Risser, P, Maruoka, M, Mao, W, Foster, J, Kelley, R, Pan, G, Gurney, A.L, de Vos, A.M, Starovasnik, M.A. | | Deposit date: | 2001-08-03 | | Release date: | 2001-09-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | IL-17s adopt a cystine knot fold: structure and activity of a novel cytokine, IL-17F, and implications for receptor binding.
EMBO J., 20, 2001
|
|
3ZMT
 
 | | LSD1-CoREST in complex with PRSFLV peptide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, PEPTIDE, ... | | Authors: | Tortorici, M, Borrello, M.T, Tardugno, M, Chiarelli, L.R, Pilotto, S, Ciossani, G, Vellore, N.A, Cowan, J, O'Connell, M, Mai, A, Baron, R, Ganesan, A, Mattevi, A. | | Deposit date: | 2013-02-12 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Protein Recognition by Small Peptide Reversible Inhibitors of the Chromatin-Modifying Lsd1/Corest Lysine Demethylase.
Acs Chem.Biol., 8, 2013
|
|
6FE8
 
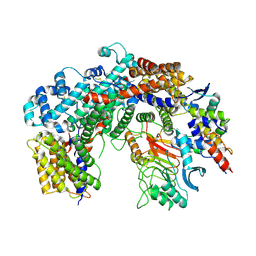 | | Cryo-EM structure of the core Centromere Binding Factor 3 complex | | Descriptor: | Centromere DNA-binding protein complex CBF3 subunit B, Centromere DNA-binding protein complex CBF3 subunit C, Suppressor of kinetochore protein 1 | | Authors: | Zhang, W.J, Lukoynova, N, Miah, S, Vaughan, C.K. | | Deposit date: | 2017-12-30 | | Release date: | 2018-08-01 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Insights into Centromere DNA Bending Revealed by the Cryo-EM Structure of the Core Centromere Binding Factor 3 with Ndc10.
Cell Rep, 24, 2018
|
|
3CB6
 
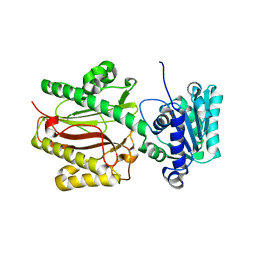 | | Crystal Structure of the S. pombe Peptidase Homology Domain of FACT complex subunit Spt16 (form B) | | Descriptor: | FACT complex subunit spt16 | | Authors: | Stuwe, T, Hothorn, M, Lejeune, E, Bortfeld-Miller, M, Scheffzek, K, Ladurner, A.G. | | Deposit date: | 2008-02-21 | | Release date: | 2008-06-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The FACT Spt16 "peptidase" domain is a histone H3-H4 binding module
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
1JSV
 
 | |
1JT4
 
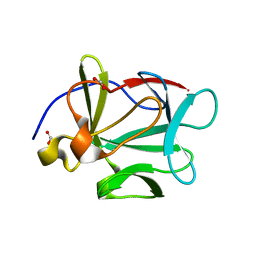 | | Human Acidic Fibroblast Growth Factor. 141 Amino Acid Form with Amino Terminal His Tag AND VAL 109 REPLACED BY LEU (V109L) | | Descriptor: | FORMIC ACID, acidic fibroblast growth factor | | Authors: | Brych, S.R, Blaber, S.I, Logan, T.M, Blaber, M. | | Deposit date: | 2001-08-20 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure and stability effects of mutations designed to increase the primary sequence symmetry within the core
region of a beta-trefoil.
Protein Sci., 10, 2001
|
|
1JT7
 
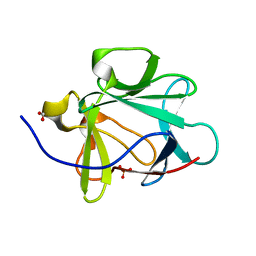 | | Human Acidic Fibroblast Growth Factor. 141 Amino Acid Form with Amino Terminal His Tag AND LEU 44 REPLACED BY PHE AND LEU 73 REPLACED BY VAL AND VAL 109 REPLACED BY LEU (L44F/L73V/V109L) | | Descriptor: | FORMIC ACID, SULFATE ION, acidic fibroblast growth factor | | Authors: | Brych, S.R, Blaber, S.I, Logan, T.M, Blaber, M. | | Deposit date: | 2001-08-20 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and stability effects of mutations designed to increase the primary sequence symmetry within the core
region of a beta-trefoil.
Protein Sci., 10, 2001
|
|
1JGG
 
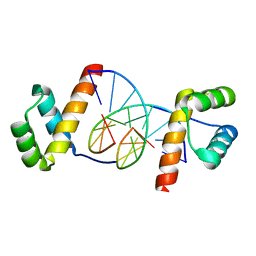 | | Even-skipped Homeodomain Complexed to AT-rich DNA | | Descriptor: | 5'-D(P*AP*AP*TP*TP*CP*AP*AP*TP*TP*A)-3', 5'-D(P*TP*AP*AP*TP*TP*GP*AP*AP*TP*T)-3', Segmentation Protein Even-Skipped | | Authors: | Hirsch, J.A, Aggarwal, A.K. | | Deposit date: | 2001-06-25 | | Release date: | 2001-07-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the even-skipped homeodomain complexed to AT-rich DNA: new perspectives on homeodomain specificity.
EMBO J., 14, 1995
|
|
2ZQS
 
 | |
3ULE
 
 | |
3UNK
 
 | | CDK2 in complex with inhibitor YL5-083 | | Descriptor: | 4-({4-[(2-chlorophenyl)amino]pyrimidin-2-yl}amino)benzoic acid, Cyclin-dependent kinase 2, PHOSPHATE ION | | Authors: | Zhu, J.-Y, Martin, M.P, Alam, R, Schonbrunn, E. | | Deposit date: | 2011-11-15 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Novel Mechanism by Which Small Molecule Inhibitors Induce the DFG Flip in Aurora A.
Acs Chem.Biol., 7, 2012
|
|
3KAB
 
 | | Structure-guided design of alpha-amino acid-derived Pin1 inhibitors | | Descriptor: | 6-methyl-1H-indole-2-carboxylic acid, DODECAETHYLENE GLYCOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, ... | | Authors: | Baker, L.M, Dokurno, P, Robinson, D.A, Surgenor, A.E, Murray, J.B, Potter, A.J, Moore, J.D. | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structure-guided design of alpha-amino acid-derived Pin1 inhibitors
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2ZQV
 
 | |
