5E6T
 
 | |
5WJ6
 
 | | Crystal structure of glutaminase C in complex with inhibitor 2-phenyl-N-{5-[4-({5-[(phenylacetyl)amino]-1,3,4-thiadiazol-2-yl}amino)piperidin-1-yl]-1,3,4-thiadiazol-2-yl}acetamide (UPGL-00004) | | Descriptor: | 2-phenyl-N-{5-[4-({5-[(phenylacetyl)amino]-1,3,4-thiadiazol-2-yl}amino)piperidin-1-yl]-1,3,4-thiadiazol-2-yl}acetamide, Glutaminase kidney isoform, mitochondrial | | Authors: | Huang, Q, Cerione, R.A. | | Deposit date: | 2017-07-21 | | Release date: | 2018-01-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.445 Å) | | Cite: | Characterization of the interactions of potent allosteric inhibitors with glutaminase C, a key enzyme in cancer cell glutamine metabolism.
J. Biol. Chem., 293, 2018
|
|
5ZJU
 
 | |
1PLG
 
 | | EVIDENCE FOR THE EXTENDED HELICAL NATURE OF POLYSACCHARIDE EPITOPES. THE 2.8 ANGSTROMS RESOLUTION STRUCTURE AND THERMODYNAMICS OF LIGAND BINDING OF AN ANTIGEN BINDING FRAGMENT SPECIFIC FOR ALPHA-(2->8)-POLYSIALIC ACID | | Descriptor: | IGG2A=KAPPA= | | Authors: | Evans, S.V, Sigurskjold, B.W, Jennings, H.J, Brisson, J.-R, Tse, W.C, To, R, Altman, E, Frosch, M, Weisgerber, C, Kratzin, H, Klebert, S, Vaesen, M, Bitter-Suermann, D, Rose, D.R, Young, N.M, Bundle, D.R. | | Deposit date: | 1995-04-24 | | Release date: | 1996-04-03 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Evidence for the extended helical nature of polysaccharide epitopes. The 2.8 A resolution structure and thermodynamics of ligand binding of an antigen binding fragment specific for alpha-(2-->8)-polysialic acid.
Biochemistry, 34, 1995
|
|
5WD7
 
 | | Structure of a bacterial polysialyltransferase in complex with fondaparinux | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-3,6-di-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-methyl 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranoside, SULFATE ION, SiaD | | Authors: | Worrall, L.J, Lizak, C, Strynadka, N.C.J. | | Deposit date: | 2017-07-04 | | Release date: | 2017-08-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | X-ray crystallographic structure of a bacterial polysialyltransferase provides insight into the biosynthesis of capsular polysialic acid.
Sci Rep, 7, 2017
|
|
5GZ0
 
 | | Crystal structure of FM329, a recombinant Fab adopted from cetuximab | | Descriptor: | FM329 heavy chain, FM329 light chain | | Authors: | Sim, D.W, Kim, J.H, Kim, Y.P, Won, H.S. | | Deposit date: | 2016-09-26 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of FM329, a recombinant Fab adopted from cetuximab
To Be Published
|
|
1EMN
 
 | | NMR STUDY OF A PAIR OF FIBRILLIN CA2+ BINDING EPIDERMAL GROWTH FACTOR-LIKE DOMAINS, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | CALCIUM ION, FIBRILLIN | | Authors: | Downing, A.K, Campbell, I.D, Handford, P.A. | | Deposit date: | 1996-08-05 | | Release date: | 1996-12-23 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a pair of calcium-binding epidermal growth factor-like domains: implications for the Marfan syndrome and other genetic disorders.
Cell(Cambridge,Mass.), 85, 1996
|
|
5G5X
 
 | |
1EMO
 
 | | NMR STUDY OF A PAIR OF FIBRILLIN CA2+ BINDING EPIDERMAL GROWTH FACTOR-LIKE DOMAINS, 22 STRUCTURES | | Descriptor: | CALCIUM ION, FIBRILLIN | | Authors: | Downing, A.K, Campbell, I.D, Handford, P.A. | | Deposit date: | 1996-08-05 | | Release date: | 1996-12-23 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a pair of calcium-binding epidermal growth factor-like domains: implications for the Marfan syndrome and other genetic disorders.
Cell(Cambridge,Mass.), 85, 1996
|
|
6JOD
 
 | | Angiotensin II type 2 receptor with ligand | | Descriptor: | Angiotensin II, Heavy chain of 4A03Fab, Light chain of 4A03Fab, ... | | Authors: | Asada, H, Iwata, S, Hirata, K, Shimamura, T. | | Deposit date: | 2019-03-20 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Crystal Structure of Angiotensin II Type 2 Receptor with Endogenous Peptide Hormone.
Structure, 28, 2020
|
|
1RMD
 
 | | RAG1 DIMERIZATION DOMAIN | | Descriptor: | RAG1, ZINC ION | | Authors: | Bellon, S.F, Rodgers, K.K, Schatz, D.G, Coleman, J.E, Steitz, T.A. | | Deposit date: | 1997-01-10 | | Release date: | 1997-07-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the RAG1 dimerization domain reveals multiple zinc-binding motifs including a novel zinc binuclear cluster.
Nat.Struct.Biol., 4, 1997
|
|
5WCN
 
 | |
8QZ3
 
 | | Crystal structure of human two pore domain potassium ion channel TREK-2 (K2P10.1) in complex with an activatory nanobody (Nb67) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Nanobody 67, POTASSIUM ION, ... | | Authors: | Baronina, A, Pike, A.C.W, Rodstrom, K.E.J, Ang, J, Bushell, S.R, Chalk, R, Mukhopadhyay, S.M.M, Pardon, E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Burgess-Brown, N.A, Tucker, S.J, Steyaert, J, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-26 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Extracellular modulation of TREK-2 activity with nanobodies provides insight into the mechanisms of K2P channel regulation.
Nat Commun, 15, 2024
|
|
4ZH7
 
 | | Structural basis of Lewisb antigen binding by the Helicobacter pylori adhesin BabA | | Descriptor: | Outer membrane protein-adhesin, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose | | Authors: | Howard, T, Hage, N, Phillips, C, Brassington, C.A, Debreczeni, J, Overman, R, Gellert, P, Stolnik, S, Winkler, G.S, Falcone, F.H. | | Deposit date: | 2015-04-24 | | Release date: | 2015-08-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural basis of Lewis(b) antigen binding by the Helicobacter pylori adhesin BabA.
Sci Adv, 1, 2015
|
|
1TCR
 
 | | MURINE T-CELL ANTIGEN RECEPTOR 2C CLONE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Garcia, K.C, Degano, M, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 1996-09-12 | | Release date: | 1997-03-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An alphabeta T cell receptor structure at 2.5 A and its orientation in the TCR-MHC complex.
Science, 274, 1996
|
|
5EUK
 
 | | Cetuximab Fab in complex with F3H meditope variant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cetuximab Fab heavy chain, Cetuximab Fab light chain, ... | | Authors: | Bzymek, K.P, Williams, J.C. | | Deposit date: | 2015-11-18 | | Release date: | 2016-10-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Natural and non-natural amino-acid side-chain substitutions: affinity and diffraction studies of meditope-Fab complexes.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
5WT9
 
 | | Complex structure of PD-1 and nivolumab-Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of Nivolumab, Light Chain of Nivolumab, ... | | Authors: | Tan, S, Zhang, H, Chai, Y, Song, H, Tong, Z, Wang, Q, Qi, J, Wong, G, Zhu, X, Liu, W.J, Gao, S, Wang, Z, Shi, Y, Yang, F, Gao, G.F, Yan, J. | | Deposit date: | 2016-12-10 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | An unexpected N-terminal loop in PD-1 dominates binding by nivolumab.
Nat Commun, 8, 2017
|
|
6KMC
 
 | | Crystal structure of a Streptococcal protein G B1 mutant | | Descriptor: | ACETIC ACID, Immunoglobulin G-binding protein G B1 | | Authors: | Watanabe, H, Honda, S. | | Deposit date: | 2019-07-31 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Histidine-Mediated Intramolecular Electrostatic Repulsion for Controlling pH-Dependent Protein-Protein Interaction.
Acs Chem.Biol., 14, 2019
|
|
1T2J
 
 | |
6K2I
 
 | |
1PAJ
 
 | | NMR SOLUTION STRUCTURE AND FLEXIBILITY OF A PEPTIDE ANTIGEN REPRESENTING THE RECEPTOR BINDING DOMAIN OF PSEUDOMONAS AERUGINOSA | | Descriptor: | FIMBRIAL PROTEIN PRECURSOR, HYDROXIDE ION | | Authors: | Mcinnes, C, Sonnichsen, F.D, Kay, C.M, Hodges, R.S, Sykes, B.D. | | Deposit date: | 1993-08-25 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure and flexibility of a peptide antigen representing the receptor binding domain of Pseudomonas aeruginosa.
Biochemistry, 32, 1993
|
|
6IC5
 
 | | Human cathepsin-C in complex with dipeptidyl cyclopropyl nitrile inhibitor 2 | | Descriptor: | (2~{S})-2-azanyl-~{N}-[(1~{R},2~{R})-1-(iminomethyl)-2-[4-[4-(4-methylpiperazin-1-yl)sulfonylphenyl]phenyl]cyclopropyl]-3-thiophen-2-yl-propanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Hakansson, M, Logan, D.T, Korkmaz, B, Lesner, A, Wysocka, M, Gieldon, A, Gauthier, F, Jenne, D, Lauritzen, C, Pedersen, J. | | Deposit date: | 2018-12-02 | | Release date: | 2019-04-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design and in vivo anti-arthritic activity evaluation of a potent dipeptidyl cyclopropyl nitrile inhibitor of cathepsin C.
Biochem. Pharmacol., 164, 2019
|
|
1BWW
 
 | | BENCE-JONES IMMUNOGLOBULIN REI VARIABLE PORTION, T39K MUTANT | | Descriptor: | PROTEIN (IG KAPPA CHAIN V-I REGION REI) | | Authors: | Uson, I, Pohl, E, Schneider, T.R, Dauter, Z, Schmidt, A, Fritz, H.J, Sheldrick, G.M. | | Deposit date: | 1998-09-29 | | Release date: | 1998-10-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.7 A structure of the stabilized REIv mutant T39K. Application of local NCS restraints.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
5HVF
 
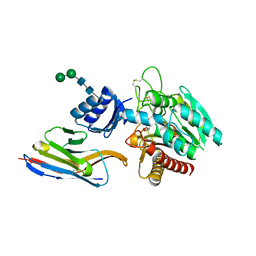 | | Crystal Structure of Thrombin-activatable Fibrinolysis Inhibitor in Complex with an Inhibitory Nanobody (VHH-i83) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CITRATE ANION, Carboxypeptidase B2, ... | | Authors: | Zhou, X, Weeks, S.D, Strelkov, S.V, Declerck, P.J. | | Deposit date: | 2016-01-28 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Elucidation of the molecular mechanisms of two nanobodies that inhibit thrombin-activatable fibrinolysis inhibitor activation and activated thrombin-activatable fibrinolysis inhibitor activity.
J.Thromb.Haemost., 14, 2016
|
|
6A48
 
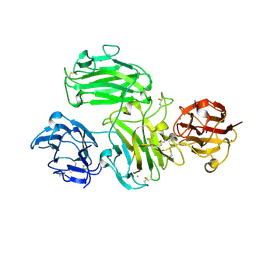 | | Crystal structure of reelin N-terminal region | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Reelin | | Authors: | Nagae, M, Takagi, J. | | Deposit date: | 2018-06-19 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural studies of reelin N-terminal region provides insights into a unique structural arrangement and functional multimerization.
J.Biochem., 169, 2021
|
|
