1RC5
 
 | |
7PKO
 
 | | CryoEM structure of Rotavirus NSP2 | | Descriptor: | Non-structural protein 2 | | Authors: | Bravo, J.P.K, Borodavka, A. | | Deposit date: | 2021-08-26 | | Release date: | 2021-09-29 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of rotavirus RNA chaperone displacement and RNA annealing.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7PKP
 
 | | NSP2 RNP complex | | Descriptor: | Non-structural protein 2 | | Authors: | Bravo, J.P.K, Borodavka, A. | | Deposit date: | 2021-08-26 | | Release date: | 2021-09-29 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of rotavirus RNA chaperone displacement and RNA annealing.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7PT0
 
 | | SCO3201 with putative ligand | | Descriptor: | SPERMIDINE, TetR family transcriptional regulator | | Authors: | Werten, S, Palm, G.J, Hinrichs, W. | | Deposit date: | 2021-09-25 | | Release date: | 2021-10-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structures of free and ligand-bound forms of the TetR/AcrR-like regulator SCO3201 from Streptomyces coelicolor suggest a novel allosteric mechanism.
Febs J., 290, 2023
|
|
2YWP
 
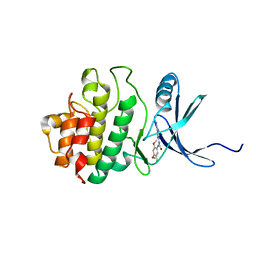 | | Crystal Structure of CHK1 with a Urea Inhibitor | | Descriptor: | 1-(5-CHLORO-2,4-DIMETHOXYPHENYL)-3-(5-CYANOPYRAZIN-2-YL)UREA, Serine/threonine-protein kinase Chk1 | | Authors: | Park, C. | | Deposit date: | 2007-04-21 | | Release date: | 2007-05-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Synthesis and biological evaluation of 1-(2,4,5-trisubstituted phenyl)-3-(5-cyanopyrazin-2-yl)ureas as potent Chk1 kinase inhibitors
Bioorg.Med.Chem.Lett., 16, 2006
|
|
5LA0
 
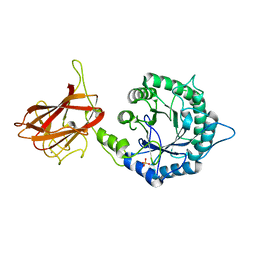 | | The mechanism by which arabinoxylanases can recognise highly decorated xylans | | Descriptor: | CALCIUM ION, Carbohydrate binding family 6, SULFATE ION, ... | | Authors: | Basle, A, Labourel, A, Cuskin, F, Jackson, A, Crouch, L, Rogowski, A, Gilbert, A. | | Deposit date: | 2016-06-13 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Mechanism by Which Arabinoxylanases Can Recognize Highly Decorated Xylans.
J.Biol.Chem., 291, 2016
|
|
2YXG
 
 | |
4K92
 
 | |
2YDI
 
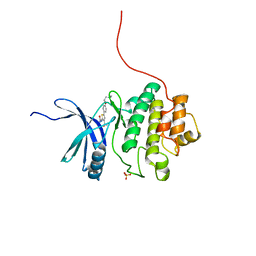 | | Discovery of Checkpoint Kinase Inhibitor AZD7762 by Structure Based Design and Optimization of Thiophene Carboxamide Ureas | | Descriptor: | 5-[4-(2-DIMETHYLAMINOETHYLOXY)PHENYL]-2-UREIDO-THIOPHENE-3-CARBOXAMIDE, SERINE/THREONINE-PROTEIN KINASE CHK1, SULFATE ION | | Authors: | Read, J.A, Breed, J, Haye, H, McCall, E, Rowsell, S, Vallentine, A, White, A. | | Deposit date: | 2011-03-21 | | Release date: | 2012-04-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of Checkpoint Kinase Inhibitor (S)-5-(3-Fluorophenyl)-N-(Piperidin-3-Yl)-3-Ureidothiophene-2-Carboxamide (Azd7762) by Structure-Based Design and Optimization of Thiophenecarboxamide Ureas.
J.Med.Chem., 55, 2012
|
|
1RMO
 
 | | Probing the Role of Tryptophans in Aequorea Victoria Green Fluorescent Proteins with an Expanded Genetic Code | | Descriptor: | wunen-nonfunctional GFP fusion protein | | Authors: | Budisa, N, Pal, P.P, Alefelder, S, Birle, P, Krywcun, T, Rubini, M, Wenger, W, Bae, J.H, Steiner, T. | | Deposit date: | 2003-11-28 | | Release date: | 2004-06-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Probing the role of tryptophans in Aequorea victoria green fluorescent proteins with an expanded genetic code
Biol.Chem., 385, 2004
|
|
3VOY
 
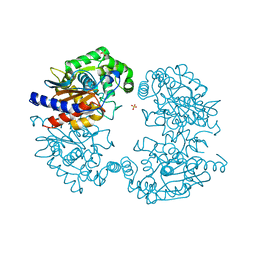 | | Crystal structure of human glutaminase in apo form | | Descriptor: | Glutaminase kidney isoform, mitochondrial, SULFATE ION | | Authors: | Thangavelu, K, Sivaraman, J. | | Deposit date: | 2012-02-23 | | Release date: | 2012-06-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the allosteric inhibitory mechanism of human kidney-type glutaminase (KGA) and its regulation by Raf-Mek-Erk signaling in cancer cell metabolism.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4KRD
 
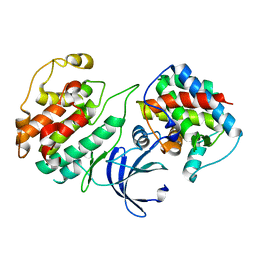 | | Crystal Structure of Pho85-Pcl10 Complex | | Descriptor: | Cyclin-dependent protein kinase PHO85, PHO85 cyclin-10 | | Authors: | Quiocho, F.A, Zheng, F. | | Deposit date: | 2013-05-16 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | New Structural Insights into Phosphorylation-free Mechanism for Full Cyclin-dependent Kinase (CDK)-Cyclin Activity and Substrate Recognition.
J.Biol.Chem., 288, 2013
|
|
1RTL
 
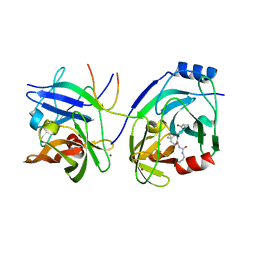 | | CRYSTAL STRUCTURE OF HCV NS3 PROTEASE DOMAIN: NS4A PEPTIDE COMPLEX WITH COVALENTLY BOUND PYRROLIDINE-5,5-TRANSLACTAM INHIBITOR | | Descriptor: | N-[(2R,3S)-1-((2S)-2-{[(CYCLOPENTYLAMINO)CARBONYL]AMINO}-3-METHYLBUTANOYL)-2-(1-FORMYL-1-CYCLOBUTYL)PYRROLIDINYL]CYCLOPROPANECARBOXAMIDE, NS3 protease/helicase, NS4A COFACTOR, ... | | Authors: | Skarzynski, T, Somers, D.O.N. | | Deposit date: | 2003-12-10 | | Release date: | 2004-12-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Pyrrolidine-5,5-trans-lactams. 4. Incorporation of a P3/P4 urea leads to potent intracellular inhibitors of hepatitis C virus NS3/4A protease
Org.Lett., 5, 2003
|
|
1RMM
 
 | | Probing the Role of Tryptophans in Aequorea Victoria Green Fluorescent Proteins with an Expanded Genetic Code | | Descriptor: | SIGF1-GFP fusion protein | | Authors: | Budisa, N, Pal, P.P, Alefelder, S, Birle, P, Krywcun, T, Rubini, M, Wenger, W, Bae, J.H, Steiner, T. | | Deposit date: | 2003-11-28 | | Release date: | 2004-06-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the role of tryptophans in Aequorea victoria green fluorescent proteins with an expanded genetic code
Biol.Chem., 385, 2004
|
|
3VT0
 
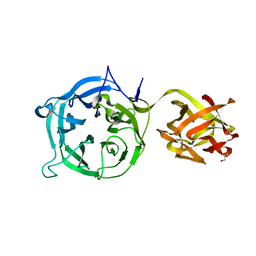 | | Crystal structure of Ct1,3Gal43A in complex with lactose | | Descriptor: | GLYCEROL, Ricin B lectin, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Jiang, D, Fan, J, Wang, X, Zhao, Y, Huang, B, Zhang, X.C. | | Deposit date: | 2012-05-18 | | Release date: | 2012-12-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.913 Å) | | Cite: | Crystal structure of 1,3Gal43A, an exo-beta-1,3-galactanase from Clostridium thermocellum
J.Struct.Biol., 180, 2012
|
|
5LSZ
 
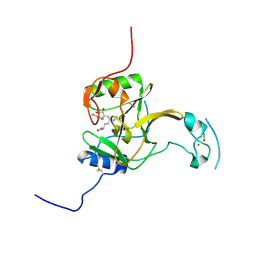 | | Structure of the Epigenetic Oncogene MMSET and inhibition by N-Alkyl Sinefungin Derivatives | | Descriptor: | Histone-lysine N-methyltransferase SETD2, THIOCYANATE ION, ZINC ION, ... | | Authors: | Tisi, D, Pathuri, P, Heightman, T. | | Deposit date: | 2016-09-05 | | Release date: | 2016-10-05 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure of the Epigenetic Oncogene MMSET and Inhibition by N-Alkyl Sinefungin Derivatives.
ACS Chem. Biol., 11, 2016
|
|
1R3F
 
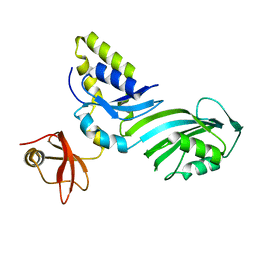 | | Crystal Structure of tRNA Pseudouridine Synthase TruB and Its RNA Complex: RNA-protein Recognition Through a Combination of Rigid Docking and Induced Fit | | Descriptor: | tRNA pseudouridine synthase B | | Authors: | Pan, H, Agarwalla, S, Moustakas, D.T, Finer-Moore, J, Stroud, R.M. | | Deposit date: | 2003-10-01 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of tRNA Pseudouridine Synthase TruB and Its RNA Complex: RNA Recognition Through a Combination of Rigid Docking and Induced Fit
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1RH8
 
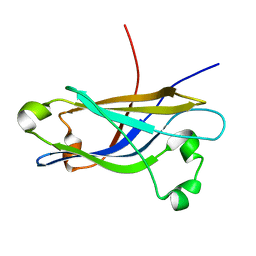 | | Three-dimensional structure of the calcium-free Piccolo C2A-domain | | Descriptor: | Piccolo protein | | Authors: | Garcia, J, Gerber, S.H, Sugita, S, Sudhof, T.C, Rizo, J. | | Deposit date: | 2003-11-14 | | Release date: | 2004-01-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A conformational switch in the Piccolo C2A domain regulated by alternative splicing.
Nat.Struct.Mol.Biol., 11, 2004
|
|
3VP3
 
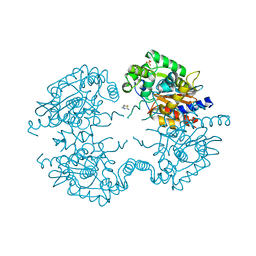 | | Crystal structure of human glutaminase in complex with inhibitor 3 | | Descriptor: | 5,5'-pentane-1,5-diylbis(1,3,4-thiadiazol-2-amine), Glutaminase kidney isoform, mitochondrial, ... | | Authors: | Thangavelu, K, Sivaraman, J. | | Deposit date: | 2012-02-23 | | Release date: | 2012-06-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the allosteric inhibitory mechanism of human kidney-type glutaminase (KGA) and its regulation by Raf-Mek-Erk signaling in cancer cell metabolism.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3VP1
 
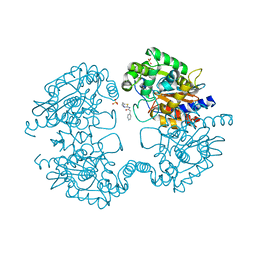 | |
3VP4
 
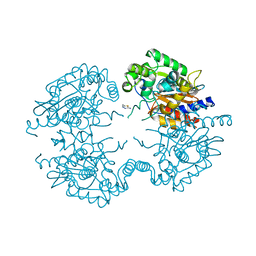 | | Crystal structure of human glutaminase in complex with inhibitor 4 | | Descriptor: | 5,5'-butane-1,4-diylbis(1,3,4-thiadiazol-2-amine), Glutaminase kidney isoform, mitochondrial | | Authors: | Thangavelu, K, Sivaraman, J. | | Deposit date: | 2012-02-23 | | Release date: | 2012-06-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis for the allosteric inhibitory mechanism of human kidney-type glutaminase (KGA) and its regulation by Raf-Mek-Erk signaling in cancer cell metabolism.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3VSZ
 
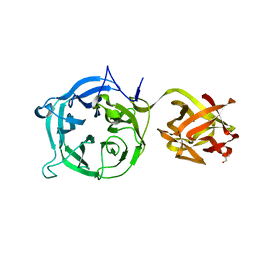 | | Crystal structure of Ct1,3Gal43A in complex with galactan | | Descriptor: | GLYCEROL, Ricin B lectin, beta-D-galactopyranose-(1-3)-beta-D-galactopyranose, ... | | Authors: | Jiang, D, Fan, J, Wang, X, Zhao, Y, Huang, B, Zhang, X.C. | | Deposit date: | 2012-05-18 | | Release date: | 2012-12-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.893 Å) | | Cite: | Crystal structure of 1,3Gal43A, an exo-beta-1,3-galactanase from Clostridium thermocellum
J.Struct.Biol., 180, 2012
|
|
3WMY
 
 | | Crystal Structure of Streptomyces coelicolor alpha-L-arabinofuranosidase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Fujimoto, Z, Maehara, T, Ichinose, H, Michikawa, M, Harazono, K, Kaneko, S. | | Deposit date: | 2013-11-29 | | Release date: | 2014-02-05 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure and characterization of the glycoside hydrolase family 62 alpha-L-arabinofuranosidase from Streptomyces coelicolor
J.Biol.Chem., 289, 2014
|
|
3WN0
 
 | | Crystal Structure of Streptomyces coelicolor alpha-L-arabinofuranosidase in complex with L-arabinose | | Descriptor: | CALCIUM ION, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Fujimoto, Z, Maehara, T, Ichinose, H, Michikawa, M, Harazono, K, Kaneko, S. | | Deposit date: | 2013-11-29 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and characterization of the glycoside hydrolase family 62 alpha-L-arabinofuranosidase from Streptomyces coelicolor
J.Biol.Chem., 289, 2014
|
|
3ZYH
 
 | | CRYSTAL STRUCTURE OF PA-IL LECTIN COMPLEXED WITH GALBG0 AT 1.5 A RESOLUTION | | Descriptor: | 3-(beta-D-galactopyranosylthio)propanoic acid, CALCIUM ION, PA-I galactophilic lectin | | Authors: | Kadam, R.U, Bergmann, M, Hurley, M, Garg, D, Cacciarini, M, Swiderska, M.A, Nativi, C, Sattler, M, Smyth, A.R, Williams, P, Camara, M, Stocker, A, Darbre, T, Reymond, J.-L. | | Deposit date: | 2011-08-23 | | Release date: | 2011-09-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | A Glycopeptide Dendrimer Inhibitor of the Galactose Specific Lectin Leca and of Pseudomonas Aeruginosa Biofilms
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
