3DXJ
 
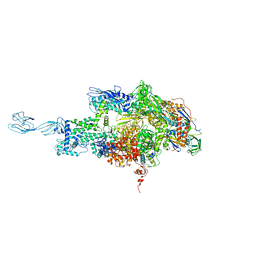 | | Crystal structure of thermus thermophilus rna polymerase holoenzyme in complex with the antibiotic myxopyronin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, BACTERIAL RNA POLYMERASE BETA SUBUNIT; CHAIN C, M, ... | | Authors: | Das, K, Arnold, E. | | Deposit date: | 2008-07-24 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The RNA polymerase "switch region" is a target for inhibitors.
Cell(Cambridge,Mass.), 135, 2008
|
|
7P0M
 
 | |
7P09
 
 | |
7P0B
 
 | |
7ZYW
 
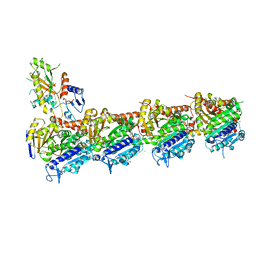 | | Crystal structure of T2R-TTL-PM534 complex | | Descriptor: | (4R)-N-[(1R)-1-[4-(cyclopropylmethoxy)-6-oxidanylidene-pyran-2-yl]butyl]-4-methyl-2-[(E)-C-methyl-N-oxidanyl-carbonimidoyl]-5H-1,3-thiazole-4-carboxamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Oliva, M.A, Diaz, J.F, Cuevas, C. | | Deposit date: | 2022-05-25 | | Release date: | 2023-11-29 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | PM534, an Optimized Target-Protein Interaction Strategy through the Colchicine Site of Tubulin.
J.Med.Chem., 67, 2024
|
|
1L9U
 
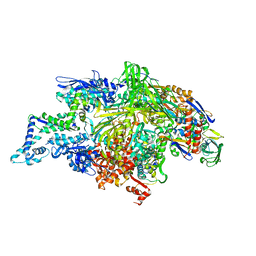 | | THERMUS AQUATICUS RNA POLYMERASE HOLOENZYME AT 4 A RESOLUTION | | Descriptor: | MAGNESIUM ION, RNA POLYMERASE, ALPHA SUBUNIT, ... | | Authors: | Murakami, K.S, Masuda, S, Darst, S.A. | | Deposit date: | 2002-03-26 | | Release date: | 2002-05-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 A resolution.
Science, 296, 2002
|
|
5NJH
 
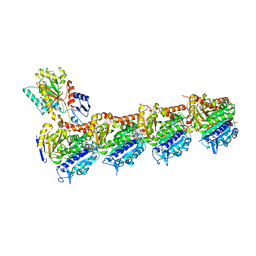 | | Triazolopyrimidines stabilize microtubules by binding to the vinca inhibitor site of tubulin | | Descriptor: | 5-chloranyl-7-[(1~{R},5~{S})-3-methoxy-8-azabicyclo[3.2.1]octan-8-yl]-6-[2,4,6-tris(fluoranyl)phenyl]-[1,2,4]triazolo[1,5-a]pyrimidine, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Sharma, A, Calvo, G.S, Prota, A.E, Diaz, J.F, Steinmetz, M.O. | | Deposit date: | 2017-03-28 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.394 Å) | | Cite: | Triazolopyrimidines Are Microtubule-Stabilizing Agents that Bind the Vinca Inhibitor Site of Tubulin.
Cell Chem Biol, 24, 2017
|
|
3MTE
 
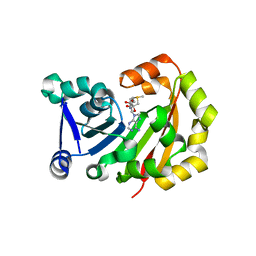 | | Crystal Structure of 16S rRNA Methyltranferase | | Descriptor: | 16S rRNA methylase, S-ADENOSYLMETHIONINE | | Authors: | Macmaster, R.A, Conn, G.L, Zelinskaya, N. | | Deposit date: | 2010-04-30 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.805 Å) | | Cite: | Structural insights into the function of aminoglycoside-resistance A1408 16S rRNA methyltransferases from antibiotic-producing and human pathogenic bacteria.
Nucleic Acids Res., 38, 2010
|
|
4GWQ
 
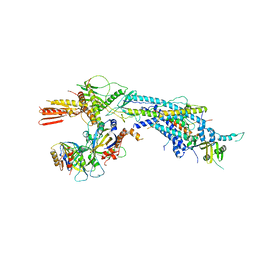 | | Structure of the Mediator Head Module from S. cerevisiae in complex with the carboxy-terminal domain (CTD) of RNA Polymerase II Rpb1 subunit | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, Mediator of RNA polymerase II transcription subunit 11, Mediator of RNA polymerase II transcription subunit 17, ... | | Authors: | Robinson, P.J.J, Bushnell, D.A, Trnka, M.J, Burlingame, A.L, Kornberg, R.D. | | Deposit date: | 2012-09-03 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Structure of the Mediator Head module bound to the carboxy-terminal domain of RNA polymerase II.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2XI5
 
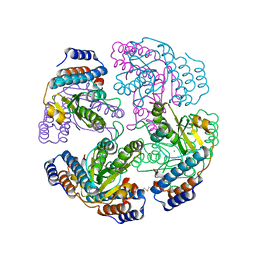 | | N-terminal endonuclease domain of La Crosse virus L-protein | | Descriptor: | MANGANESE (II) ION, RNA POLYMERASE L | | Authors: | Reguera, J, Weber, F, Cusack, S. | | Deposit date: | 2010-06-28 | | Release date: | 2010-10-06 | | Last modified: | 2012-07-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bunyaviridae RNA Polymerases (L-Protein) Have an N-Terminal, Influenza-Like Endonuclease Domain, Essential for Viral CAP-Dependent Transcription.
Plos Pathog., 6, 2010
|
|
2XI7
 
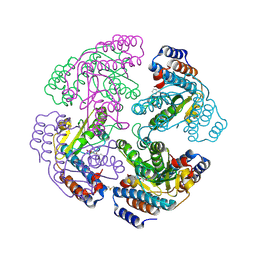 | | N-terminal endonuclease domain of La Crosse virus L-protein | | Descriptor: | 2-4-DIOXO-4-PHENYLBUTANOIC ACID, MANGANESE (II) ION, RNA POLYMERASE L | | Authors: | Reguera, J, Weber, F, Cusack, S. | | Deposit date: | 2010-06-28 | | Release date: | 2010-10-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bunyaviridae RNA Polymerases (L-Protein) Have an N-Terminal, Influenza-Like Endonuclease Domain, Essential for Viral CAP-Dependent Transcription.
Plos Pathog., 6, 2010
|
|
3ADG
 
 | |
4GGK
 
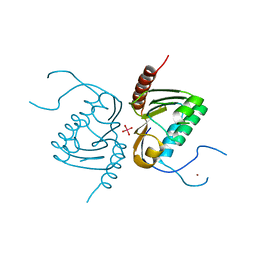 | | Crystal structure of Zucchini from mouse (mZuc / PLD6 / MitoPLD) bound to tungstate | | Descriptor: | Mitochondrial cardiolipin hydrolase, TUNGSTATE(VI)ION, ZINC ION | | Authors: | Ipsaro, J.J, Haase, A.D, Hannon, G.J, Joshua-Tor, L. | | Deposit date: | 2012-08-06 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structural biochemistry of Zucchini implicates it as a nuclease in piRNA biogenesis.
Nature, 491, 2012
|
|
7U87
 
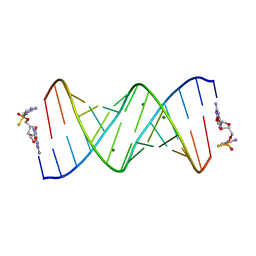 | | Product of 13mer primer with activated G monomer diastereomer 1 | | Descriptor: | 5'-O-[(R)-(2-amino-1H-imidazol-1-yl)(sulfanyl)phosphoryl]guanosine, MAGNESIUM ION, RNA (5'-R(*(LCC)P*(LCC)P*(LCC)P*(LCG)P*AP*CP*UP*UP*AP*AP*GP*UP*CP*(G46)P*(G46))-3') | | Authors: | Fang, Z, Szostak, J.W. | | Deposit date: | 2022-03-08 | | Release date: | 2023-03-15 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Catalytic Metal Ion-Substrate Coordination during Nonenzymatic RNA Primer Extension.
J.Am.Chem.Soc., 2024
|
|
7U88
 
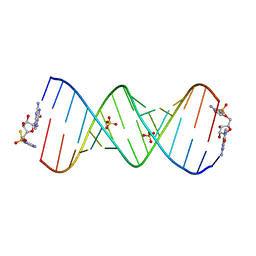 | | Product of 13mer primer with activated G monomer diastereomer 2 | | Descriptor: | 5'-O-[(R)-(2-amino-1H-imidazol-1-yl)(sulfanyl)phosphoryl]guanosine, RNA (5'-R(*(LKC)P*(LCC)P*(LCC)P*(LCG)P*AP*CP*UP*UP*AP*AP*GP*UP*CP*(G46)P*(G46))-3'), SULFATE ION | | Authors: | Fang, Z, Szostak, J.W. | | Deposit date: | 2022-03-08 | | Release date: | 2023-03-15 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Catalytic Metal Ion-Substrate Coordination during Nonenzymatic RNA Primer Extension.
J.Am.Chem.Soc., 2024
|
|
7U89
 
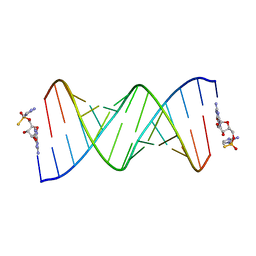 | | Product of 14mer primer with activated G monomer diastereomer 1 | | Descriptor: | 5'-O-[(R)-(2-amino-1H-imidazol-1-yl)(sulfanyl)phosphoryl]guanosine, RNA (5'-R(*(LKC)P*(LCC)P*(LCC)P*(LCG)P*AP*CP*UP*UP*AP*AP*GP*UP*CP*GP*(G46))-3') | | Authors: | Fang, Z, Szostak, J.W. | | Deposit date: | 2022-03-08 | | Release date: | 2023-03-15 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Catalytic Metal Ion-Substrate Coordination during Nonenzymatic RNA Primer Extension.
J.Am.Chem.Soc., 2024
|
|
7U8A
 
 | | Product of 14mer primer with activated G monomer diastereomer 2 | | Descriptor: | 5'-O-[(R)-(2-amino-1H-imidazol-1-yl)(sulfanyl)phosphoryl]guanosine, MAGNESIUM ION, RNA (5'-R(*(LKC)P*(LCC)P*(LCC)P*(LCG)P*AP*CP*UP*UP*AP*AP*GP*UP*CP*GP*(G46))-3') | | Authors: | Fang, Z, Szostak, J.W. | | Deposit date: | 2022-03-08 | | Release date: | 2023-03-15 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Catalytic Metal Ion-Substrate Coordination during Nonenzymatic RNA Primer Extension.
J.Am.Chem.Soc., 2024
|
|
1Y3S
 
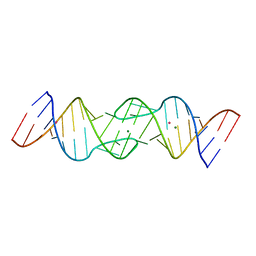 | | HIV-1 DIS RNA subtype F- MPD form | | Descriptor: | 5'-R(*CP*UP*(5BU)P*GP*CP*UP*GP*AP*AP*GP*UP*GP*CP*AP*CP*AP*CP*AP*GP*CP*AP*AP*G)-3', MAGNESIUM ION, POTASSIUM ION | | Authors: | Ennifar, E, Dumas, P. | | Deposit date: | 2004-11-26 | | Release date: | 2005-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Polymorphism of Bulged-out Residues in HIV-1 RNA DIS Kissing Complex and Structure Comparison with Solution Studies.
J.Mol.Biol., 356, 2006
|
|
488D
 
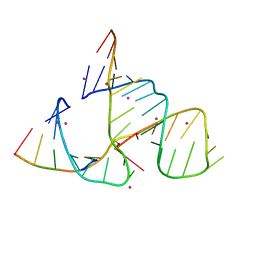 | | CATALYTIC RNA ENZYME-PRODUCT COMPLEX | | Descriptor: | CADMIUM ION, FIRST RNA FRAGMENT OF CLEAVED SUBSTRATE, RNA RIBOZYME STRAND, ... | | Authors: | Murray, J.B, Szoke, H, Szoke, A, Scott, W.G. | | Deposit date: | 2000-02-25 | | Release date: | 2000-03-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Capture and visualization of a catalytic RNA enzyme-product complex using crystal lattice trapping and X-ray holographic reconstruction.
Mol.Cell, 5, 2000
|
|
1L9Z
 
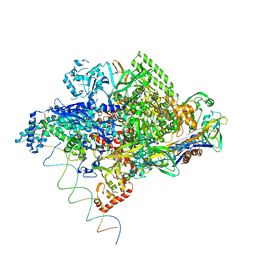 | | Thermus aquaticus RNA Polymerase Holoenzyme/Fork-Junction Promoter DNA Complex at 6.5 A Resolution | | Descriptor: | MAGNESIUM ION, RNA POLYMERASE, ALPHA SUBUNIT, ... | | Authors: | Murakami, K.S, Masuda, S, Campbell, E.A, Muzzin, O, Darst, S.A. | | Deposit date: | 2002-03-27 | | Release date: | 2002-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (6.5 Å) | | Cite: | Structural basis of transcription initiation: an RNA polymerase holoenzyme-DNA complex.
Science, 296, 2002
|
|
1A79
 
 | |
2KXN
 
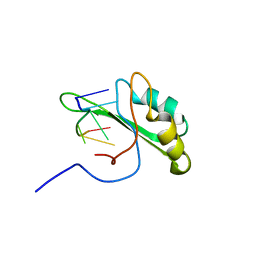 | | NMR structure of human Tra2beta1 RRM in complex with AAGAAC RNA | | Descriptor: | 5'-R(*AP*AP*GP*AP*AP*C)-3', Transformer-2 protein homolog beta | | Authors: | Clery, A, Jayne, S, Benderska, N, Dominguez, C, Stamm, S, Allain, F.H.-T. | | Deposit date: | 2010-05-10 | | Release date: | 2011-03-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular basis of purine-rich RNA recognition by the human SR-like protein Tra2-beta1
Nat.Struct.Mol.Biol., 18, 2011
|
|
2JUK
 
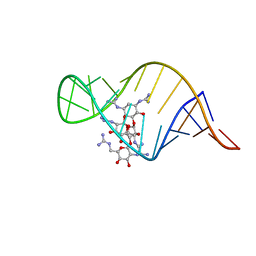 | | guanidino neomycin B recognition of an HIV-1 RNA helix | | Descriptor: | (1S,2R,3S,4R,6S)-4,6-bis{[amino(iminio)methyl]amino}-2-{[3-O-(2,6-bis{[amino(iminio)methyl]amino}-2,6-dideoxy-beta-L-glucopyranosyl)-beta-D-arabinofuranosyl]oxy}-3-hydroxycyclohexyl 2,6-bis{[amino(iminio)methyl]amino}-2,6-dideoxy-beta-L-glucopyranoside, HIV-1 frameshift site RNA | | Authors: | Staple, D.W, Venditti, V, Niccolai, N, Elson-Schwab, L, Tor, Y, Butcher, S.E. | | Deposit date: | 2007-08-30 | | Release date: | 2007-11-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Guanidinoneomycin B Recognition of an HIV-1 RNA Helix.
Chembiochem, 9, 2008
|
|
1SAQ
 
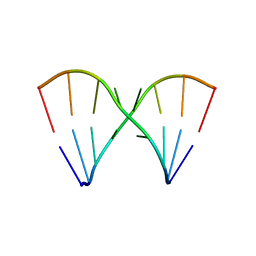 | | Crystal Structure of the RNA octamer GIC(GA)GCC | | Descriptor: | 5'-R(*GP*IP*CP*GP*AP*GP*CP*C)-3' | | Authors: | Jang, S.B, Baeyens, K, Jeong, M.S, SantaLucia Jr, J, Turner, D, Holbrook, S.R. | | Deposit date: | 2004-02-09 | | Release date: | 2004-05-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of two RNA octamers containing tandem G.A base pairs.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1ICG
 
 | | STRUCTURE OF THE HYBRID RNA/DNA R-GCUUCGGC-D[F]U IN PRESENCE OF IR(NH3)6+++ | | Descriptor: | 5'-R(*GP*CP*UP*UP*CP*GP*GP*C)-D(P*(UFP))-3', CHLORIDE ION, IRIDIUM HEXAMMINE ION | | Authors: | Cruse, W, Saludjian, P, Neuman, A, Prange, T. | | Deposit date: | 2001-03-31 | | Release date: | 2001-04-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Destabilizing effect of a fluorouracil extra base in a hybrid RNA duplex compared with bromo and chloro analogues.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
