5ZPW
 
 | | Generation of a long-acting fusion inhibitor against HIV-1 | | Descriptor: | MET-THR-TRP-GLU-GLU-TRP-ASP-MK8-LYS-ILE-GLU-MK8-TYR-THR-MK8-LYS-ILE-GLU-MK8-LEU-ILE-LYS-LYS-SER, Transmembrane protein gp41 | | Authors: | Guo, Y, Shi, X.L. | | Deposit date: | 2018-04-16 | | Release date: | 2019-03-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Generation of a long-acting fusion inhibitor against HIV-1.
Medchemcomm, 9, 2018
|
|
8JHH
 
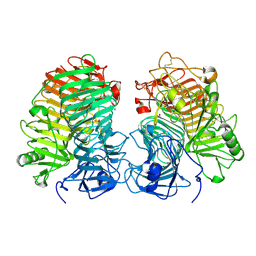 | | Glycoside hydrolase family 55 endo-beta-1,3-glucanase from Microdochium nivale | | Descriptor: | GLYCEROL, MnLam55A | | Authors: | Ota, T, Saburi, W, Yamashita, K, Tagami, T, Yu, J, Komba, S, Jewell, L.E, Hsiang, T, Imai, R, Yao, M, Mori, H. | | Deposit date: | 2023-05-23 | | Release date: | 2024-04-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular mechanism for endo-type action of glycoside hydrolase family 55 endo-beta-1,3-glucanase on beta 1-3/1-6-glucan.
J.Biol.Chem., 299, 2023
|
|
5WJ6
 
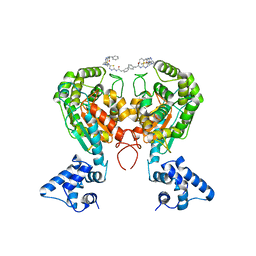 | | Crystal structure of glutaminase C in complex with inhibitor 2-phenyl-N-{5-[4-({5-[(phenylacetyl)amino]-1,3,4-thiadiazol-2-yl}amino)piperidin-1-yl]-1,3,4-thiadiazol-2-yl}acetamide (UPGL-00004) | | Descriptor: | 2-phenyl-N-{5-[4-({5-[(phenylacetyl)amino]-1,3,4-thiadiazol-2-yl}amino)piperidin-1-yl]-1,3,4-thiadiazol-2-yl}acetamide, Glutaminase kidney isoform, mitochondrial | | Authors: | Huang, Q, Cerione, R.A. | | Deposit date: | 2017-07-21 | | Release date: | 2018-01-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.445 Å) | | Cite: | Characterization of the interactions of potent allosteric inhibitors with glutaminase C, a key enzyme in cancer cell glutamine metabolism.
J. Biol. Chem., 293, 2018
|
|
5WC9
 
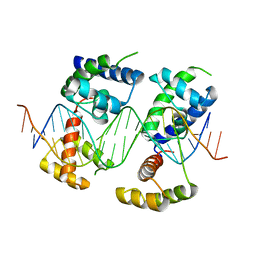 | | Human Pit-1 and 4xCATT DNA complex | | Descriptor: | DNA (5'-D(*CP*CP*AP*TP*TP*CP*AP*TP*TP*CP*AP*TP*TP*CP*AP*TP*TP*CP*GP*GP*A)-3'), DNA (5'-D(*CP*CP*GP*AP*AP*TP*GP*AP*AP*TP*GP*AP*AP*TP*GP*AP*AP*TP*GP*GP*T)-3'), Pituitary-specific positive transcription factor 1 | | Authors: | Agarwal, S, Cho, T.Y. | | Deposit date: | 2017-06-29 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Biochemical and structural characterization of a novel cooperative binding mode by Pit-1 with CATT repeats in the macrophage migration inhibitory factor promoter.
Nucleic Acids Res., 46, 2018
|
|
5IJJ
 
 | |
5MHS
 
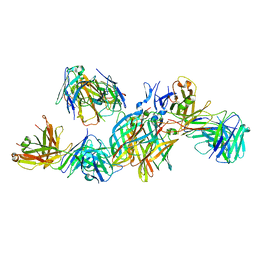 | | T1L reovirus sigma1 complexed with 5C6 Fab fragments | | Descriptor: | 5C6 Fab heavy chain, 5C6 Fab light chain, Outer capsid protein sigma-1 | | Authors: | Stehle, T, Dietrich, M.H. | | Deposit date: | 2016-11-25 | | Release date: | 2016-12-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural Insights into Reovirus sigma 1 Interactions with Two Neutralizing Antibodies.
J. Virol., 91, 2017
|
|
3KPW
 
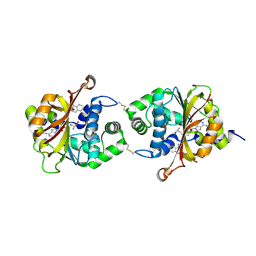 | | Crystal Structure of hPNMT in Complex AdoHcy and 1-Aminoisoquinoline | | Descriptor: | ISOQUINOLIN-1-AMINE, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Drinkwater, N, Martin, J.L. | | Deposit date: | 2009-11-17 | | Release date: | 2010-09-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Fragment-based screening by X-ray crystallography, MS and isothermal titration calorimetry to identify PNMT (phenylethanolamine N-methyltransferase) inhibitors.
Biochem.J., 431, 2010
|
|
4ZHE
 
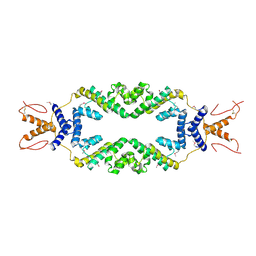 | | Crystal structure of the SeMet substituted Topless related protein 2 (TPR2) N-terminal domain (1-209) from rice | | Descriptor: | ASPR2 protein | | Authors: | Ke, J, Ma, H, Gu, X, Brunzelle, J.S, Xu, H.E, Melcher, K. | | Deposit date: | 2015-04-24 | | Release date: | 2015-08-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for recognition of diverse transcriptional repressors by the TOPLESS family of corepressors.
Sci Adv, 1, 2015
|
|
5K0R
 
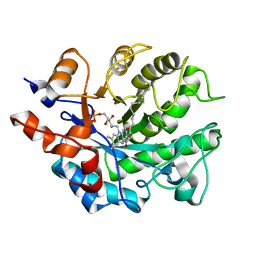 | | Crystal structure of reduced Shewanella Yellow Enzyme 4 (SYE4) | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, NAD(P)H:flavin oxidoreductase Sye4, Octadecane | | Authors: | Elegheert, J, Brige, A, Savvides, S.N. | | Deposit date: | 2016-05-17 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural dissection of Shewanella oneidensis old yellow enzyme 4 bound to a Meisenheimer complex and (nitro)phenolic ligands.
FEBS Lett., 591, 2017
|
|
4ZFB
 
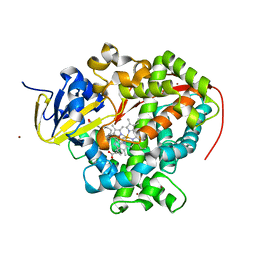 | | Cytochrome P450 pentamutant from BM3 bound to Palmitic Acid | | Descriptor: | 1,2-ETHANEDIOL, Bifunctional P-450/NADPH-P450 reductase, NICKEL (II) ION, ... | | Authors: | Rogers, W.E, Othman, T, Heidary, D.K, Huxford, T. | | Deposit date: | 2015-04-21 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Effect of Mutation and Substrate Binding on the Stability of Cytochrome P450BM3 Variants.
Biochemistry, 55, 2016
|
|
6H1O
 
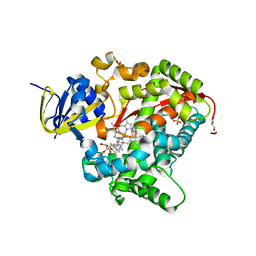 | | Structure of the BM3 heme domain in complex with voriconazole | | Descriptor: | 1,2-ETHANEDIOL, Bifunctional cytochrome P450/NADPH--P450 reductase, GLYCEROL, ... | | Authors: | Jeffreys, L.N, Munro, A.W.M, Leys, D. | | Deposit date: | 2018-07-12 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.734 Å) | | Cite: | Novel insights into P450 BM3 interactions with FDA-approved antifungal azole drugs.
Sci Rep, 9, 2019
|
|
6H6Z
 
 | |
6QC5
 
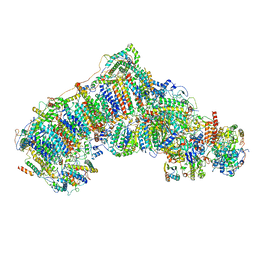 | | Ovine respiratory complex I FRC closed class 1 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Letts, J.A, Sazanov, L.A. | | Deposit date: | 2018-12-26 | | Release date: | 2019-08-21 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structures of Respiratory Supercomplex I+III2Reveal Functional and Conformational Crosstalk.
Mol.Cell, 75, 2019
|
|
9J1U
 
 | |
4ZFA
 
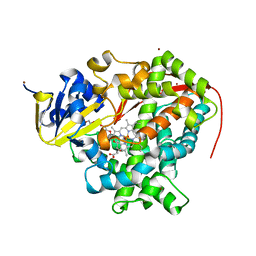 | | Cytochrome P450 wild type from BM3 with bound PEG | | Descriptor: | 1,2-ETHANEDIOL, Bifunctional P-450/NADPH-P450 reductase, NICKEL (II) ION, ... | | Authors: | Rogers, W.E, Othman, T, Heidary, D.K, Huxford, T. | | Deposit date: | 2015-04-21 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.765 Å) | | Cite: | Effect of Mutation and Substrate Binding on the Stability of Cytochrome P450BM3 Variants.
Biochemistry, 55, 2016
|
|
5WV3
 
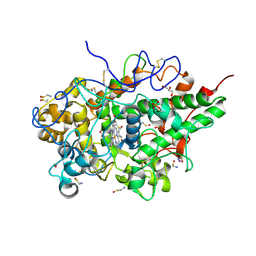 | | Crystal structure of bovine lactoperoxidase with a partial Glu258-heme linkage at 2.07 A resolution. | | Descriptor: | 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Singh, P.K, Sirohi, H.V, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2016-12-21 | | Release date: | 2017-02-15 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural basis of activation of mammalian heme peroxidases
Prog. Biophys. Mol. Biol., 133, 2018
|
|
3KKS
 
 | |
4ZF6
 
 | | Cytochrome P450 pentamutant from BM3 with bound PEG | | Descriptor: | 1,2-ETHANEDIOL, Bifunctional P-450/NADPH-P450 reductase, NICKEL (II) ION, ... | | Authors: | Rogers, W.E, Othman, T, Heidary, D.K, Huxford, T. | | Deposit date: | 2015-04-21 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.773 Å) | | Cite: | Effect of Mutation and Substrate Binding on the Stability of Cytochrome P450BM3 Variants.
Biochemistry, 55, 2016
|
|
6T8Y
 
 | | NAD+-dependent fungal formate dehydrogenase from Chaetomium thermophilum: A complex with the reduced form of the cofactor NADH and the substrate formate at a secondary site. | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Isupov, M.N, Yelmazer, B, De Rose, S.A, Littlechild, J.A. | | Deposit date: | 2019-10-25 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Structural insights into the NAD + -dependent formate dehydrogenase mechanism revealed from the NADH complex and the formate NAD + ternary complex of the Chaetomium thermophilum enzyme.
J.Struct.Biol., 212, 2020
|
|
5N2F
 
 | | Structure of PD-L1/small-molecule inhibitor complex | | Descriptor: | 4-[[4-[[3-(2,3-dihydro-1,4-benzodioxin-6-yl)-2-methyl-phenyl]methoxy]-2,5-bis(fluoranyl)phenyl]methylamino]-3-oxidanylidene-butanoic acid, Programmed cell death 1 ligand 1 | | Authors: | Guzik, K, Zak, K.M, Grudnik, P, Dubin, G, Holak, T.A. | | Deposit date: | 2017-02-07 | | Release date: | 2017-06-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Small-Molecule Inhibitors of the Programmed Cell Death-1/Programmed Death-Ligand 1 (PD-1/PD-L1) Interaction via Transiently Induced Protein States and Dimerization of PD-L1.
J. Med. Chem., 60, 2017
|
|
9KU6
 
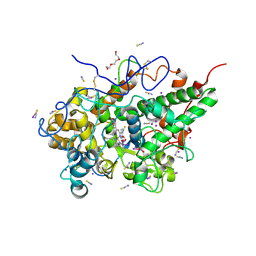 | | Crystal structure of the complex of lactoperoxidase with nitric oxide at 1.72 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Maurya, A, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2024-12-03 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structure of the complex of lactoperoxidase with nitric oxide at 1.72 A resolution
To Be Published
|
|
5K8K
 
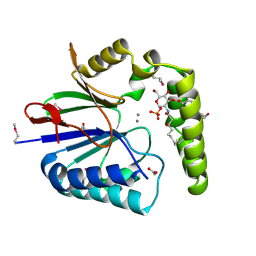 | | Structure of the Haemophilus influenzae LpxH-lipid X complex | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, ACETATE ION, GLYCEROL, ... | | Authors: | Cho, J, Lee, C.-J, Zhou, P. | | Deposit date: | 2016-05-30 | | Release date: | 2016-08-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of the essential Haemophilus influenzae UDP-diacylglucosamine pyrophosphohydrolase LpxH in lipid A biosynthesis.
Nat Microbiol, 1, 2016
|
|
5XD0
 
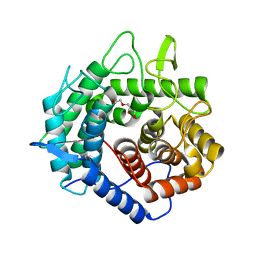 | | Apo Structure of Beta-1,3-1,4-glucanase from Paenibacillus sp.X4 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Glucanase, TRIETHYLENE GLYCOL | | Authors: | Baek, S.C, Ho, T.-H, Kang, L.-W, Kim, H. | | Deposit date: | 2017-03-24 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Improvement of enzyme activity of beta-1,3-1,4-glucanase from Paenibacillus sp. X4 by error-prone PCR and structural insights of mutated residues.
Appl. Microbiol. Biotechnol., 101, 2017
|
|
5KA6
 
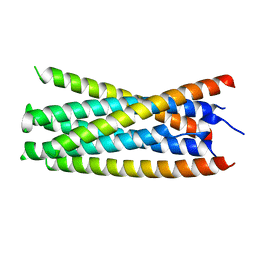 | |
3LLS
 
 | |
