6FVG
 
 | | The Structure of CK2alpha with CCh507 bound | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Casein kinase II subunit alpha, [1-[2-(phenylsulfonylamino)ethyl]piperidin-4-yl]methyl 1~{H}-indole-3-carboxylate | | Authors: | Brear, P, Prudent, R, Laudet, B, Filhol, O, Cochet, C, Sautel, C, Moucadel, V, Bestgen, B, Engel, M, Ettaoussi, M, Lomberget, T, Le Borgne, M, Kufareva, I, Abagyan, R, Hyvonen, M. | | Deposit date: | 2018-03-02 | | Release date: | 2019-06-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of holoenzyme-disrupting chemicals as substrate-selective CK2 inhibitors.
Sci Rep, 9, 2019
|
|
6FT7
 
 | | Crystal structure of CLK3 in complex with compound 8a | | Descriptor: | 1,2-ETHANEDIOL, 3-phenyl-1~{H}-pyrrolo[3,4-g]indol-8-one, Dual specificity protein kinase CLK3, ... | | Authors: | Chaikuad, A, Walter, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Kunick, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-20 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Molecular structures of cdc2-like kinases in complex with a new inhibitor chemotype.
PLoS ONE, 13, 2018
|
|
6FTZ
 
 | | COMPLEMENT FACTOR D COMPLEXED WITH COMPOUND 6 | | Descriptor: | Complement factor D, ~{N}4-[3-(aminomethyl)phenyl]-1~{H}-indole-2,4-dicarboxamide | | Authors: | Ostermann, N. | | Deposit date: | 2018-02-26 | | Release date: | 2018-06-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Discovery and Design of First Benzylamine-Based Ligands Binding to an Unlocked Conformation of the Complement Factor D.
ACS Med Chem Lett, 9, 2018
|
|
8H55
 
 | | The structure of zebrafish angiotensinogen | | Descriptor: | Angiotensinogen, SULFATE ION | | Authors: | Zhou, A, Wei, H. | | Deposit date: | 2022-10-12 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | BIOLOGICAL IMPLICATIONS OF A 2 ANGSTROMS STRUCTURE OF DIMERIC ANGIOTENSINOGEN
To Be Published
|
|
6FUH
 
 | | Complement factor D in complex with the inhibitor (4-((3-(aminomethyl)phenyl)amino)quinazolin-2-yl)-L-valine | | Descriptor: | (2~{S})-2-[[4-[[3-(aminomethyl)phenyl]amino]quinazolin-2-yl]amino]-3-methyl-butanoic acid, Complement factor D | | Authors: | Mac Sweeney, A, Ostermann, N, Vulpetti, A, Maibaum, J, Erbel, P, Lorthiois, E, Yoon, T, Randl, S, Ruedisser, S. | | Deposit date: | 2018-02-27 | | Release date: | 2018-06-06 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Discovery and Design of First Benzylamine-Based Ligands Binding to an Unlocked Conformation of the Complement Factor D.
ACS Med Chem Lett, 9, 2018
|
|
6FYL
 
 | | X-ray structure of CLK2-KD(136-496)/CX-4945 at 1.95A | | Descriptor: | 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, Dual specificity protein kinase CLK2 | | Authors: | Kallen, J. | | Deposit date: | 2018-03-12 | | Release date: | 2018-07-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | X-ray Structures and Feasibility Assessment of CLK2 Inhibitors for Phelan-McDermid Syndrome.
ChemMedChem, 13, 2018
|
|
6G01
 
 | |
6FMA
 
 | | Crystal structure of ERK2 in complex with an adenosine derivative | | Descriptor: | 3-[6-azanyl-9-[(2~{R},3~{R},4~{S},5~{R})-5-(azidomethyl)-3,4-bis(oxidanyl)oxolan-2-yl]purin-8-yl]sulfanylpropanoic acid, DIMETHYL SULFOXIDE, Mitogen-activated protein kinase 1, ... | | Authors: | Gelin, M, Labesse, G. | | Deposit date: | 2018-01-30 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.667 Å) | | Cite: | None
To be published
|
|
6FMO
 
 | |
6FN8
 
 | | Domain II of the HUMAN COPPER CHAPERONE FOR SUPEROXIDE DISMUTASE at 1.55 A resolution | | Descriptor: | Copper chaperone for superoxide dismutase, ZINC ION | | Authors: | Sala, F.A, Wright, G.S.A, Antonyuk, S.V, Garratt, R.C, Hasnain, S.S. | | Deposit date: | 2018-02-02 | | Release date: | 2019-01-30 | | Last modified: | 2019-06-19 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Molecular recognition and maturation of SOD1 by its evolutionarily destabilised cognate chaperone hCCS.
Plos Biol., 17, 2019
|
|
6G5C
 
 | | Interaction of TiO2 nanoparticles (NM-101) with Lysozyme | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Sbirkova-Dimitrova, H.I, Radoslavov, G, Hristov, P, Ganev, V, Dimowa, L, Rusev, R, Shivachev, B.L. | | Deposit date: | 2018-03-29 | | Release date: | 2018-10-24 | | Last modified: | 2019-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallization and crystal structure of lysozyme in the presence of nanosized Titanium dioxide
Bul.Chem.Commn., 50, 2018
|
|
6G61
 
 | |
6G97
 
 | |
6G9H
 
 | |
6G93
 
 | |
6G9J
 
 | |
8H4X
 
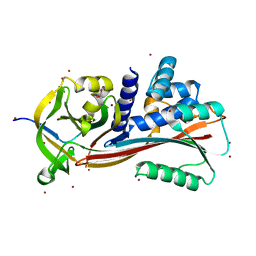 | | Structure of cleaved Serpin B9 | | Descriptor: | 1,2-ETHANEDIOL, Serpin B9, ZINC ION | | Authors: | Zhou, A, Yan, T. | | Deposit date: | 2022-10-11 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Expression, purification and crystallization of cleaved Serpin B9
To Be Published
|
|
6FR1
 
 | |
8H8V
 
 | |
8H8U
 
 | |
8H8T
 
 | |
8H7P
 
 | | Crystal structure of aqualigase bound with Suc-AAPF | | Descriptor: | 5,6-DIHYDRO-BENZO[H]CINNOLIN-3-YLAMINE, CALCIUM ION, Subtilisin, ... | | Authors: | Li, H, Ma, M.Z, Zhang, L.J, Dai, L, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-10-20 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of aqualigase bound with Suc-AAPF
To Be Published
|
|
6FX5
 
 | | MITF dimerization mutant | | Descriptor: | Microphthalmia-associated transcription factor, SULFATE ION | | Authors: | Pogenberg, V, Milewski, M, Wilmanns, M. | | Deposit date: | 2018-03-08 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Mechanism of conditional partner selectivity in MITF/TFE family transcription factors with a conserved coiled coil stammer motif.
Nucleic Acids Res., 48, 2020
|
|
1GKT
 
 | | Neutron Laue diffraction structure of endothiapepsin complexed with transition state analogue inhibitor H261 | | Descriptor: | ENDOTHIAPEPSIN, INHIBITOR, H261 | | Authors: | Coates, L, Erskine, P.T, Wood, S.P, Myles, D.A.A, Cooper, J.B. | | Deposit date: | 2001-08-20 | | Release date: | 2001-11-20 | | Last modified: | 2023-11-15 | | Method: | NEUTRON DIFFRACTION (2.1 Å) | | Cite: | A Neutron Laue Diffraction Study of Endothiapepsin: Implications for the Aspartic Proteinase Mechanism
Biochemistry, 40, 2001
|
|
1GJN
 
 | | Hydrogen Peroxide Derived Myoglobin Compound II at pH 5.2 | | Descriptor: | HYDROXIDE ION, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Hersleth, H.-P, Dalhus, B, Gorbitz, C.H, Andersson, K.K. | | Deposit date: | 2001-07-27 | | Release date: | 2002-03-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | An Iron Hydroxide Moiety in the 1.35 A Resolution Structure of Hydrogen Peroxide Derived Myoglobin Compound II at Ph 5.2
J.Biol.Inorg.Chem., 7, 2002
|
|
