5MHL
 
 | |
1B9Y
 
 | |
8D9G
 
 | | gRAMP-TPR-CHAT Non match PFS target RNA(Craspase) | | Descriptor: | CHAT domain protein, RAMP superfamily protein, RNA (36-MER), ... | | Authors: | Hu, C, Nam, K.H, Schuler, G, Ke, A. | | Deposit date: | 2022-06-09 | | Release date: | 2023-06-14 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Craspase is a CRISPR RNA-guided, RNA-activated protease.
Science, 377, 2022
|
|
7WZ4
 
 | | Structure of an orphan GPCR-G protein signaling complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Xu, J, Chen, G, Liu, Z, Du, Y. | | Deposit date: | 2022-02-17 | | Release date: | 2022-10-26 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Activation and allosteric regulation of the orphan GPR88-Gi1 signaling complex.
Nat Commun, 13, 2022
|
|
7WYB
 
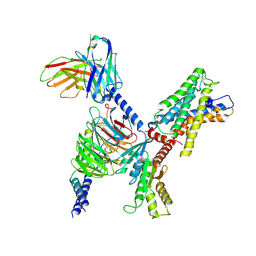 | | ADGRL3/Gi complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | He, Y, Qian, Y. | | Deposit date: | 2022-02-15 | | Release date: | 2022-10-26 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structural insights into adhesion GPCR ADGRL3 activation and G q , G s , G i , and G 12 coupling.
Mol.Cell, 82, 2022
|
|
8GUT
 
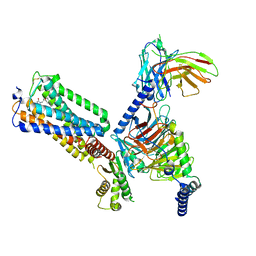 | | Cryo-EM structure of LEI-CB2-Gi complex | | Descriptor: | 1-[[4-[5-fluoranyl-6-[(oxan-4-ylamino)methyl]pyridin-2-yl]phenyl]methyl]-3-(2-methylpropyl)imidazolidine-2,4-dione, Cannabinoid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, Z.J, Hua, T, Li, X.T, Chang, H, Wu, L.J. | | Deposit date: | 2022-09-13 | | Release date: | 2023-05-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural basis of selective cannabinoid CB 2 receptor activation.
Nat Commun, 14, 2023
|
|
5LDS
 
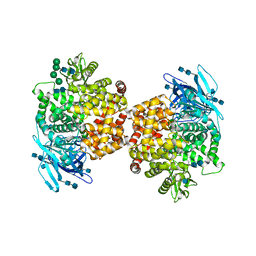 | | Structure of the porcine aminopeptidase N ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Santiago, C, Reguera, J, Mudgal, G, Casasnovas, J.M. | | Deposit date: | 2016-06-27 | | Release date: | 2017-04-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Allosteric inhibition of aminopeptidase N functions related to tumor growth and virus infection.
Sci Rep, 7, 2017
|
|
8GUR
 
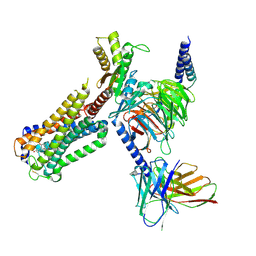 | | Cryo-EM structure of CP-CB2-G protein complex | | Descriptor: | 2-[(1R,2R,5R)-5-hydroxy-2-(3-hydroxypropyl)cyclohexyl]-5-(2-methyloctan-2-yl)phenol, Cannabinoid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Wu, L.J, Hua, T, Liu, Z.J, Li, X.T, Chang, H. | | Deposit date: | 2022-09-13 | | Release date: | 2023-05-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structural basis of selective cannabinoid CB 2 receptor activation.
Nat Commun, 14, 2023
|
|
7WY5
 
 | | ADGRL3/Gq complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Isoform 3 of Adhesion G protein-coupled receptor L3, ... | | Authors: | He, Y, Qian, Y. | | Deposit date: | 2022-02-15 | | Release date: | 2022-10-26 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structural insights into adhesion GPCR ADGRL3 activation and Gq, Gs, Gi, and G12 coupling.
Mol.Cell, 82, 2022
|
|
7WY8
 
 | | ADGRL3/Gs complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Isoform 3 of Adhesion G protein-coupled receptor L3, ... | | Authors: | He, Y, Qian, Y. | | Deposit date: | 2022-02-15 | | Release date: | 2022-10-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structural insights into adhesion GPCR ADGRL3 activation and Gq, Gs, Gi, and G12 coupling
Mol.Cell, 82, 2022
|
|
1ATL
 
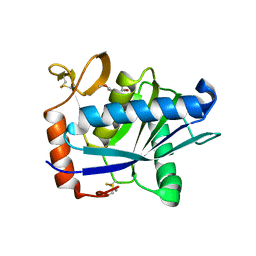 | | Structural interaction of natural and synthetic inhibitors with the VENOM METALLOPROTEINASE, ATROLYSIN C (FORM-D) | | Descriptor: | CALCIUM ION, O-methyl-N-[(2S)-4-methyl-2-(sulfanylmethyl)pentanoyl]-L-tyrosine, Snake venom metalloproteinase atrolysin-D, ... | | Authors: | Zhang, D, Botos, I, Gomis-Rueth, F.-X, Doll, R, Blood, C, Njoroge, F.G, Fox, J.W, Bode, W, Meyer, E.F. | | Deposit date: | 1995-05-26 | | Release date: | 1995-10-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural interaction of natural and synthetic inhibitors with the venom metalloproteinase, atrolysin C (form d).
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
8GW8
 
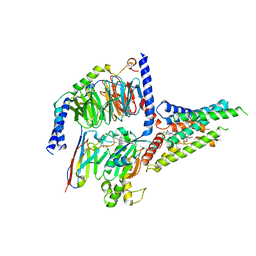 | | the human PTH1 receptor bound to an intracellular biased agonist | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Isoform Gnas-2 of Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Kobayashi, K, Kusakizako, T, Okamoto, H.H, Nureki, O. | | Deposit date: | 2022-09-16 | | Release date: | 2023-06-14 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Class B1 GPCR activation by an intracellular agonist.
Nature, 618, 2023
|
|
5T48
 
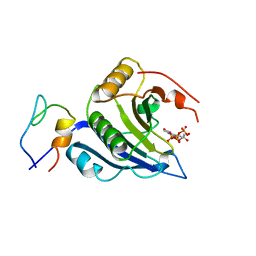 | | Crystal structure of the D. melanogaster eIF4E-eIF4G complex without lateral contact | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, Eukaryotic translation initiation factor 4E, Eukaryotic translation initiation factor 4G, ... | | Authors: | Gruener, S, Peter, D, Weber, R, Wohlbold, L, Chung, M.-Y, Weichenrieder, O, Valkov, E, Igreja, C, Izaurralde, E. | | Deposit date: | 2016-08-29 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The Structures of eIF4E-eIF4G Complexes Reveal an Extended Interface to Regulate Translation Initiation.
Mol.Cell, 64, 2016
|
|
2GBI
 
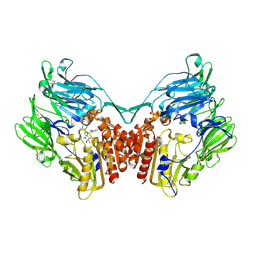 | | rat DPP-IV with xanthine inhibitor 4 | | Descriptor: | 2-({8-[(3R)-3-AMINOPIPERIDIN-1-YL]-1,3-DIMETHYL-2,6-DIOXO-1,2,3,6-TETRAHYDRO-7H-PURIN-7-YL}METHYL)BENZONITRILE, Dipeptidyl peptidase 4 | | Authors: | Longenecker, K.L, Jakob, C.G, Fry, E.H, Wilk, S. | | Deposit date: | 2006-03-10 | | Release date: | 2006-07-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal Structures of DPP-IV (CD26) from Rat Kidney Exhibit Flexible Accommodation of Peptidase-Selective Inhibitors.
Biochemistry, 45, 2006
|
|
5T8R
 
 | | Crystal structure of human BAZ2A PHD zinc finger in complex with unmodified H3 10-mer | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2A, GLYCEROL, Histone H3.1, ... | | Authors: | Amato, A, Gadd, M.S, Bortoluzzi, A, Ciulli, A. | | Deposit date: | 2016-09-08 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of molecular recognition of helical histone H3 tail by PHD finger domains.
Biochem. J., 474, 2017
|
|
4YAL
 
 | | Reduced CYPOR with 2'-AMP | | Descriptor: | ADENOSINE-2'-MONOPHOSPHATE, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Xia, C, Kim, J.J.P. | | Deposit date: | 2015-02-17 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Mutants of Cytochrome P450 Reductase Lacking Either Gly-141 or Gly-143 Destabilize Its FMN Semiquinone.
J.Biol.Chem., 291, 2016
|
|
5SWI
 
 | | Crystal structure of SpGH92 in complex with mannose | | Descriptor: | CALCIUM ION, GLYCEROL, Sugar hydrolase, ... | | Authors: | Shapiro-Ward, S, Boraston, A.B. | | Deposit date: | 2016-08-08 | | Release date: | 2016-12-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Molecular Characterization of N-glycan Degradation and Transport in Streptococcus pneumoniae and Its Contribution to Virulence.
PLoS Pathog., 13, 2017
|
|
1PEV
 
 | | Crystal Structure of the Actin Interacting Protein from Caenorhabditis Elegans | | Descriptor: | Actin interacting protein 1 | | Authors: | Vorobiev, S, Mohri, K, Fedorov, A.A, Ono, S, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-05-22 | | Release date: | 2003-07-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of functional residues on Caenorhabditis elegans actin-interacting protein 1 (UNC-78) for disassembly of actin depolymerizing factor/cofilin-bound actin filaments.
J.Biol.Chem., 279, 2004
|
|
5MMM
 
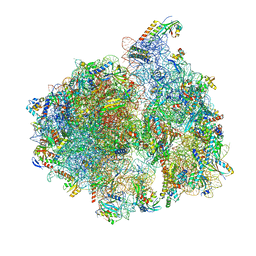 | | Structure of the 70S chloroplast ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein 2, ... | | Authors: | Bieri, P, Leibundgut, M, Saurer, M, Boehringer, D, Ban, N. | | Deposit date: | 2016-12-11 | | Release date: | 2017-01-11 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The complete structure of the chloroplast 70S ribosome in complex with translation factor pY.
EMBO J., 36, 2017
|
|
8HMV
 
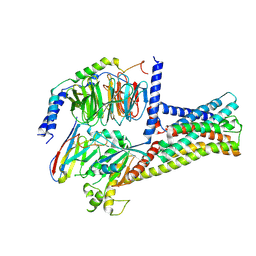 | | Structure of GPR21-Gs complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Wong, T.S, Gao, W. | | Deposit date: | 2022-12-05 | | Release date: | 2023-03-01 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Cryo-EM structure of orphan G protein-coupled receptor GPR21.
MedComm (2020), 4, 2023
|
|
2GBF
 
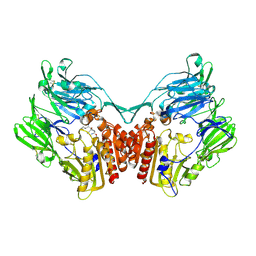 | | rat dpp-IV with alkynyl cyanopyrrolidine #1 | | Descriptor: | (1S)-2-[(2S,5R)-2-(AMINOMETHYL)-5-ETHYNYLPYRROLIDIN-1-YL]-1-CYCLOPENTYL-2-OXOETHANAMINE, Dipeptidyl peptidase 4 | | Authors: | Longenecker, K.L, Jakob, C.G, Fry, E.H, Wilk, S. | | Deposit date: | 2006-03-10 | | Release date: | 2006-07-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structures of DPP-IV (CD26) from Rat Kidney Exhibit Flexible Accommodation of Peptidase-Selective Inhibitors.
Biochemistry, 45, 2006
|
|
8HS3
 
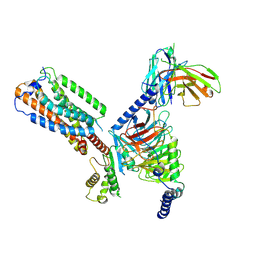 | | Gi bound orphan GPR20 in ligand-free state | | Descriptor: | Ggama, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Lin, X, Jiang, S, Xu, F. | | Deposit date: | 2022-12-16 | | Release date: | 2023-03-08 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | The activation mechanism and antibody binding mode for orphan GPR20.
Cell Discov, 9, 2023
|
|
1OC9
 
 | | TRYPAREDOXIN II FROM C.FASCICULATA solved by MR | | Descriptor: | TRYPAREDOXIN II | | Authors: | Leonard, G.A, Micossi, E, Hunter, W.N. | | Deposit date: | 2003-02-07 | | Release date: | 2003-03-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Tryparedoxins from Crithidia Fasciculata and Trypanosoma Brucei: Photoreduction of the Redox Disulfide Using Synchrotron Radiation and Evidence for a Conformational Switch Implicated in Function
J.Biol.Chem., 278, 2003
|
|
8HSC
 
 | | Gi bound Orphan GPR20 complex with Fab046 in ligand-free state | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Lin, X, Jiang, S, Xu, F. | | Deposit date: | 2022-12-19 | | Release date: | 2023-03-08 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | The activation mechanism and antibody binding mode for orphan GPR20.
Cell Discov, 9, 2023
|
|
8HJ2
 
 | | GPR21 wt with G15 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Chen, B, Lin, X, Xu, F. | | Deposit date: | 2022-11-22 | | Release date: | 2023-03-15 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of orphan GPR21 signaling complexes.
Nat Commun, 14, 2023
|
|
